Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-26 |
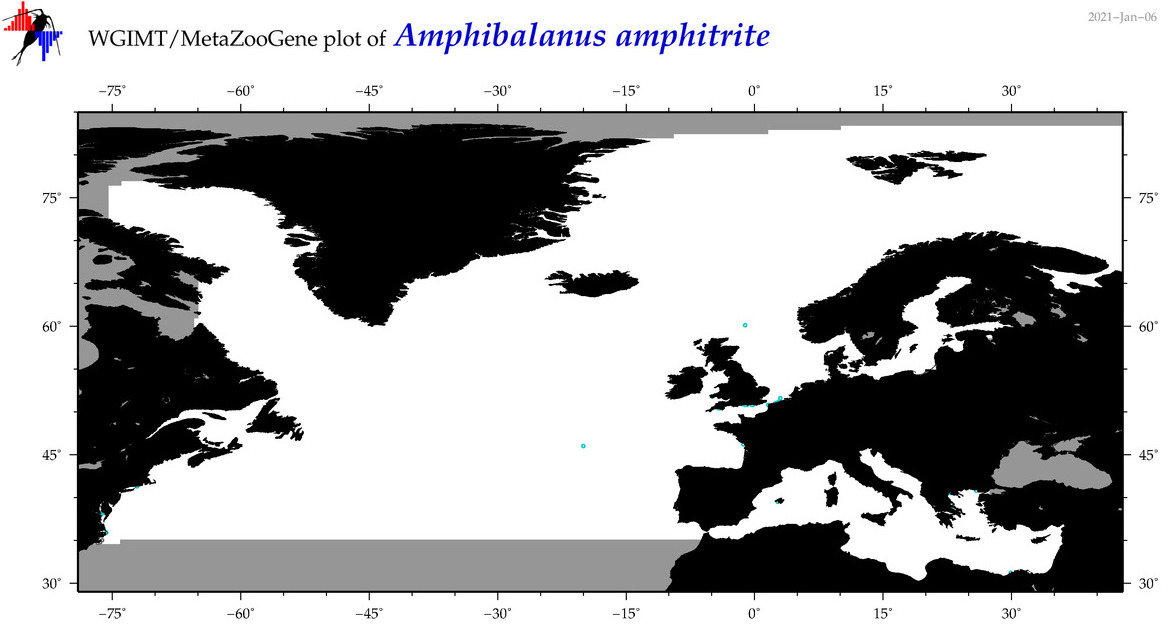
| Amphibalanus amphitrite |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 274
12S = 13
16S = 3
18S = 3
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4006760
R:1:0:0:0 |
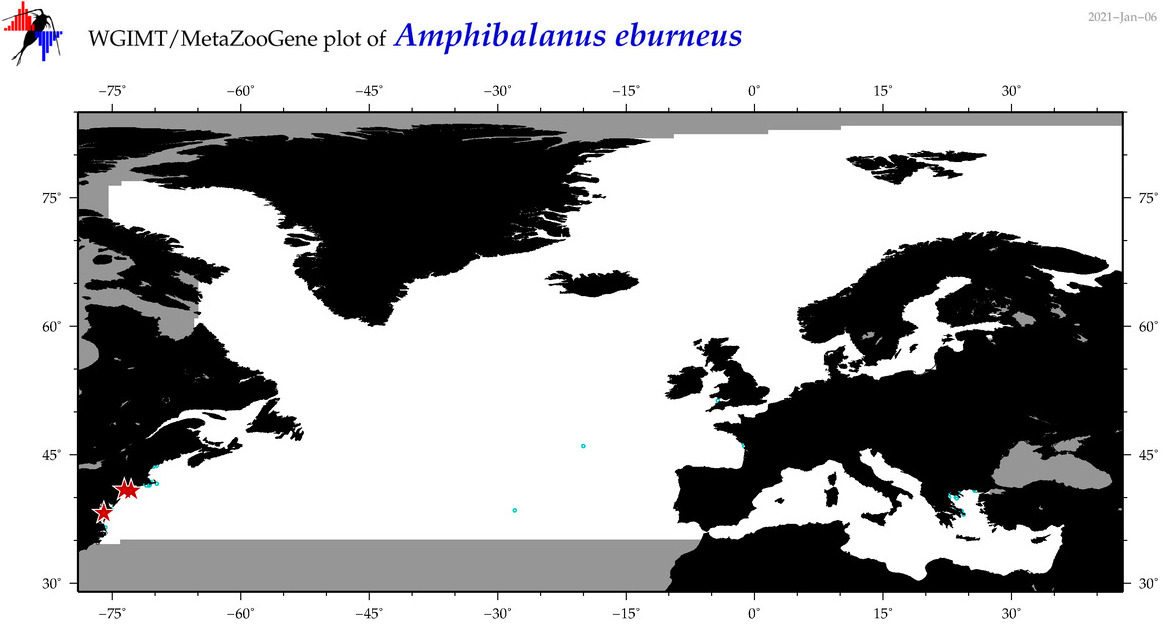
| Amphibalanus eburneus |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 74
12S = 9
16S = 1
18S = 1
28S = 1
|
COI = 14
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4006761
R:1:1:0:0 |
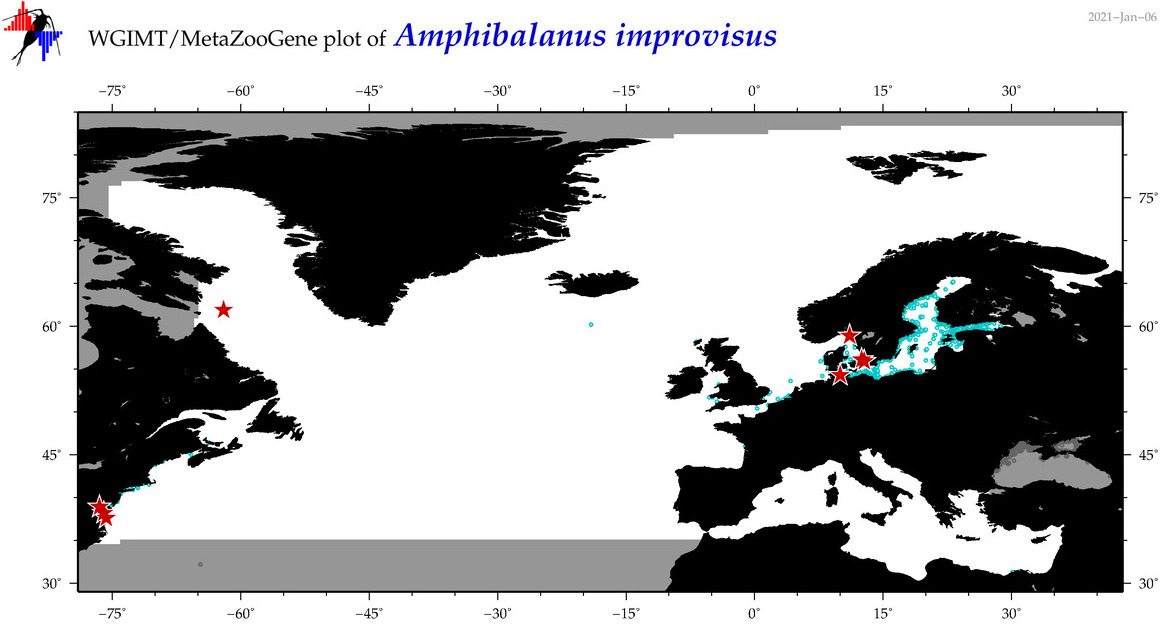
| Amphibalanus improvisus |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 59
12S = 11
16S = 8
18S = 0
28S = 0
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4003742
R:1:0:0:0 |
| Amphibalanus venustus |
Species
(4) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4043120
R:1:0:0:0 |
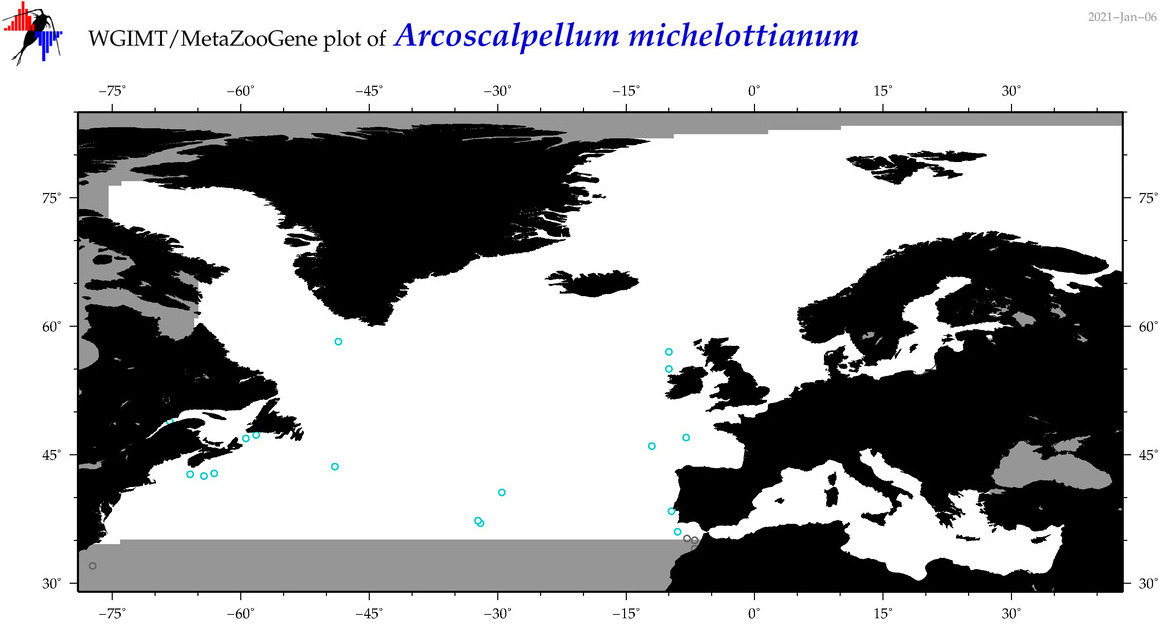
| Arcoscalpellum michelottianum |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 3
16S = 0
18S = 3
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4022777
R:1:0:0:0 |
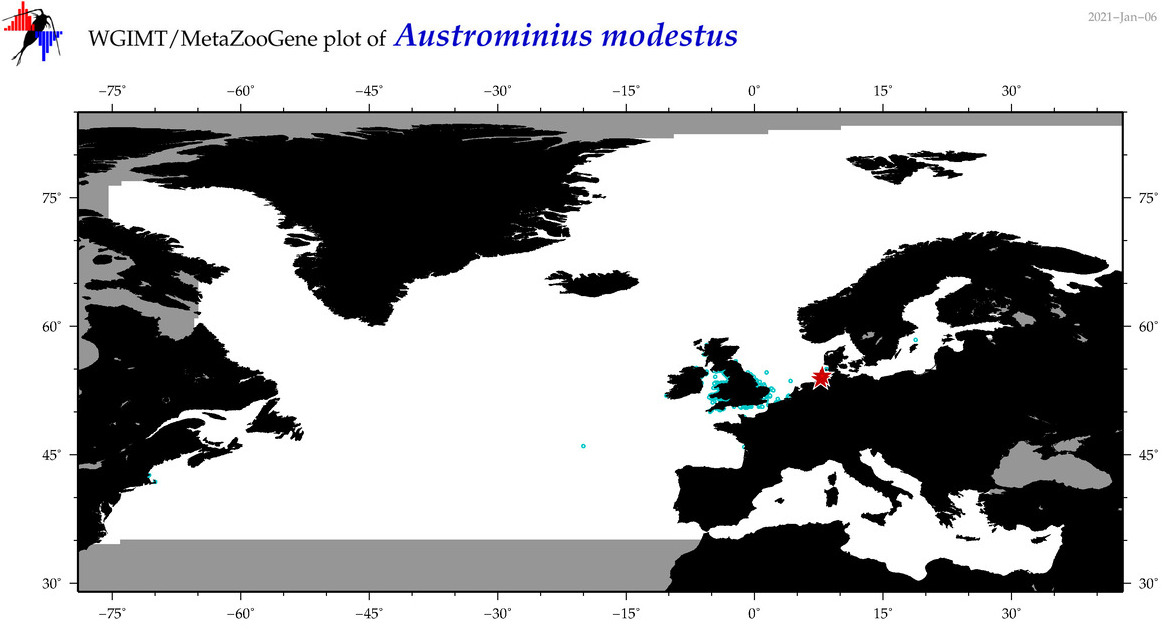
| Austrominius modestus |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 137
12S = 5
16S = 2
18S = 2
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4010240
R:1:0:0:0 |
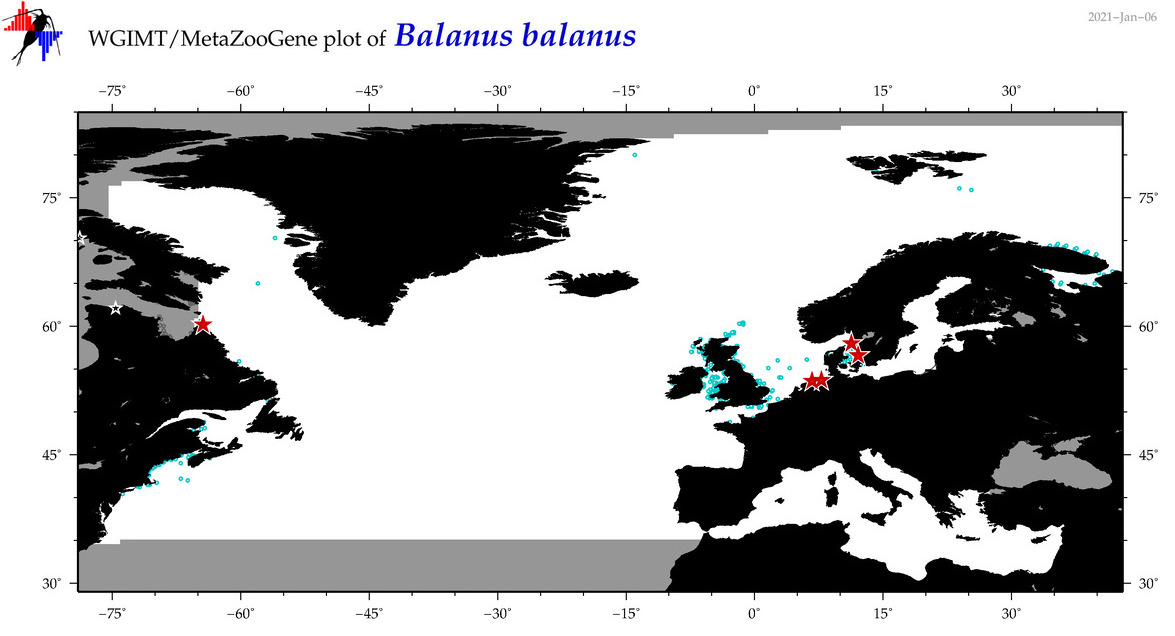
| Balanus balanus |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 38
12S = 4
16S = 3
18S = 1
28S = 1
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4000674
R:1:0:0:0 |
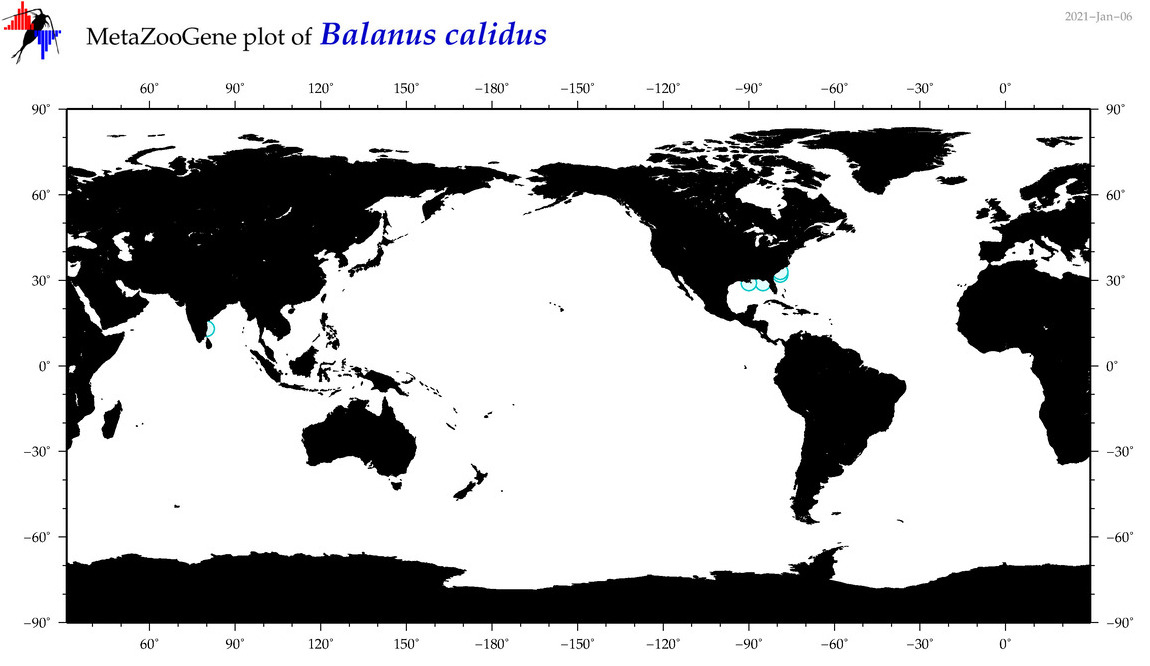
| Balanus calidus |
Species
(8) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4045084
R:1:0:0:0 |
| Balanus crenatus |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 178
12S = 2
16S = 1
18S = 2
28S = 2
|
COI = 24
12S = 0
16S = 0
18S = 0
28S = 0
|
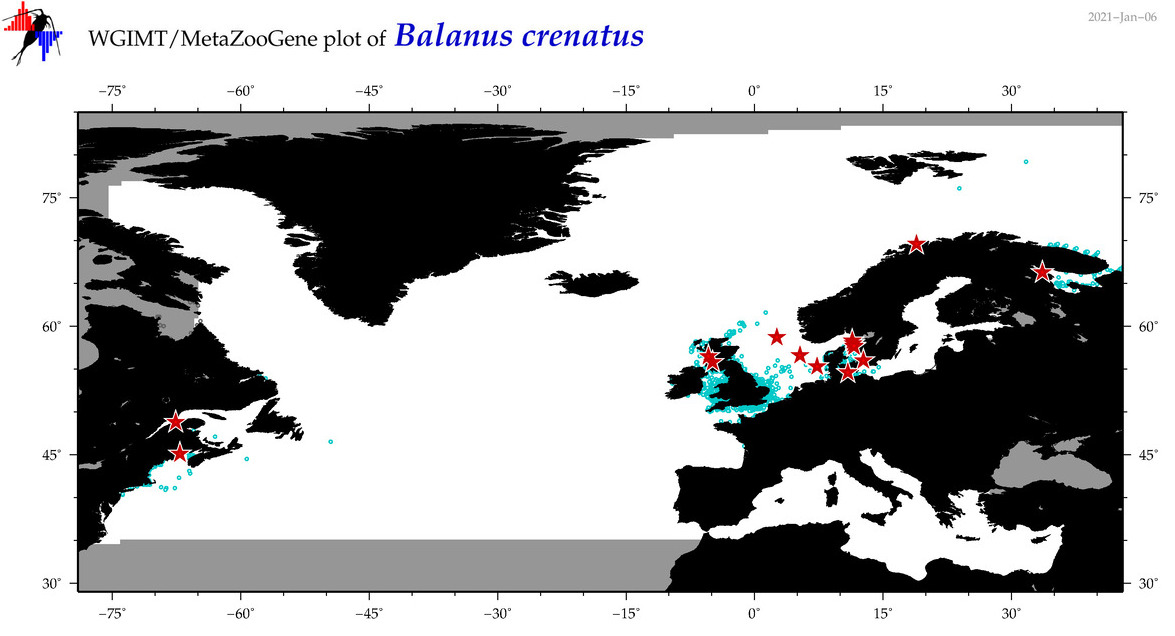 |
accepted
T4000331
R:1:0:0:0 |
| Chelonibia testudinaria |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 434
12S = 146
16S = 110
18S = 7
28S = 40
|
COI = 45
12S = 0
16S = 0
18S = 0
28S = 0
|
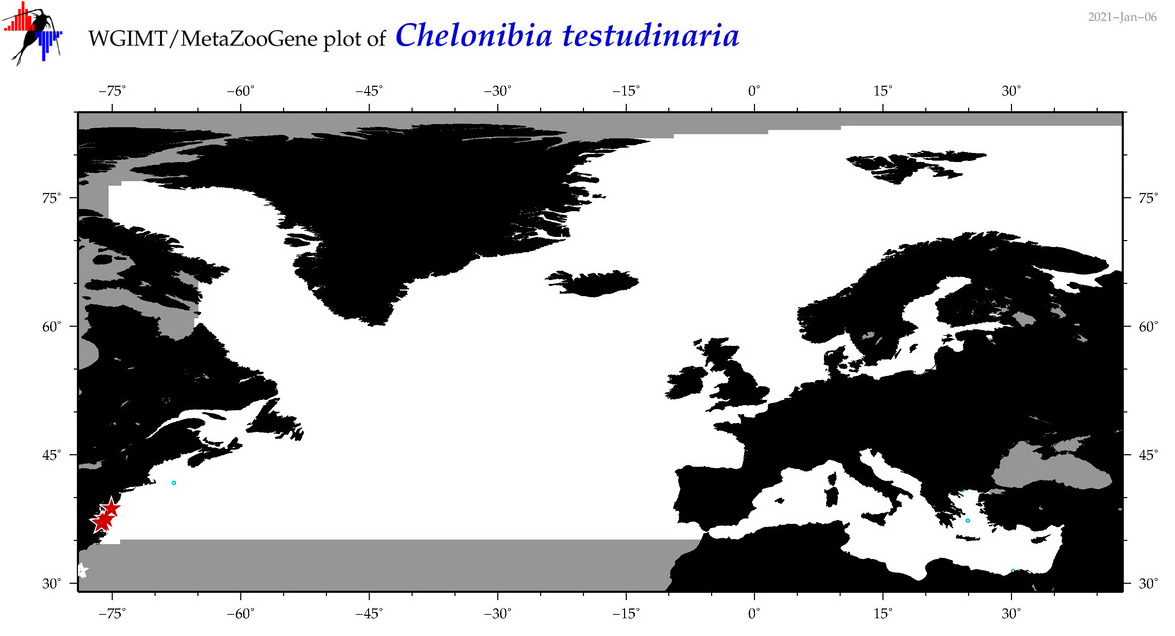 |
accepted
T4006479
R:1:0:0:0 |
| Chirona hameri |
Species
(11) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
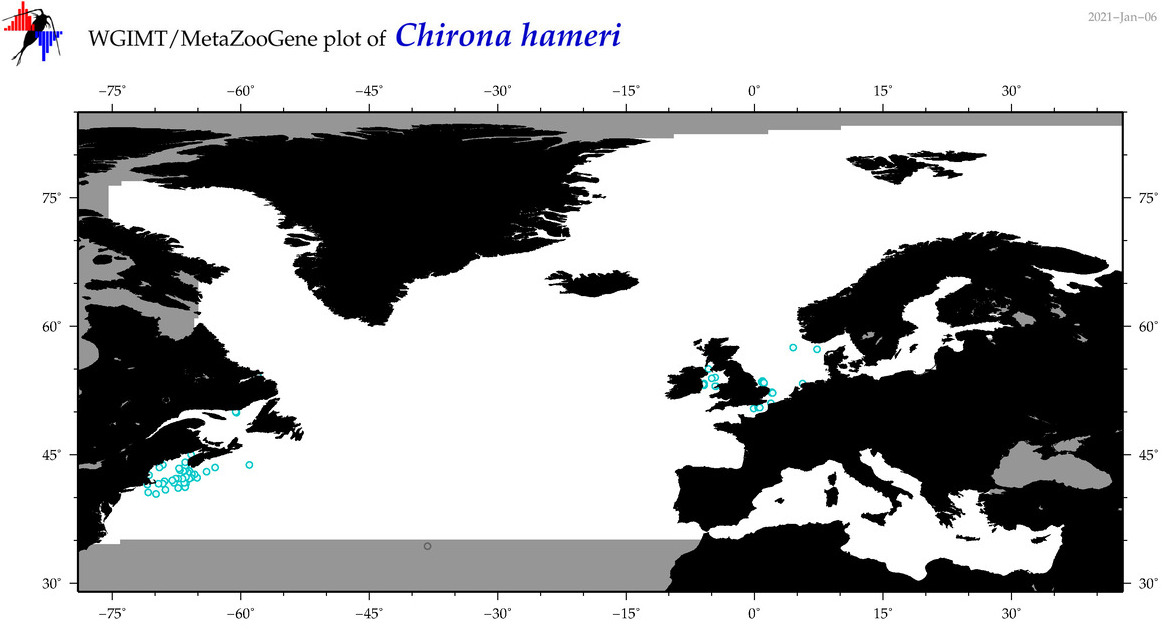 |
accepted
T4047619
R:1:0:0:0 |
| Chthamalus fragilis |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 458
12S = 1
16S = 4
18S = 2
28S = 1
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4021677
R:1:0:0:0 |
| Conchoderma virgatum |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 16
12S = 1
16S = 0
18S = 2
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
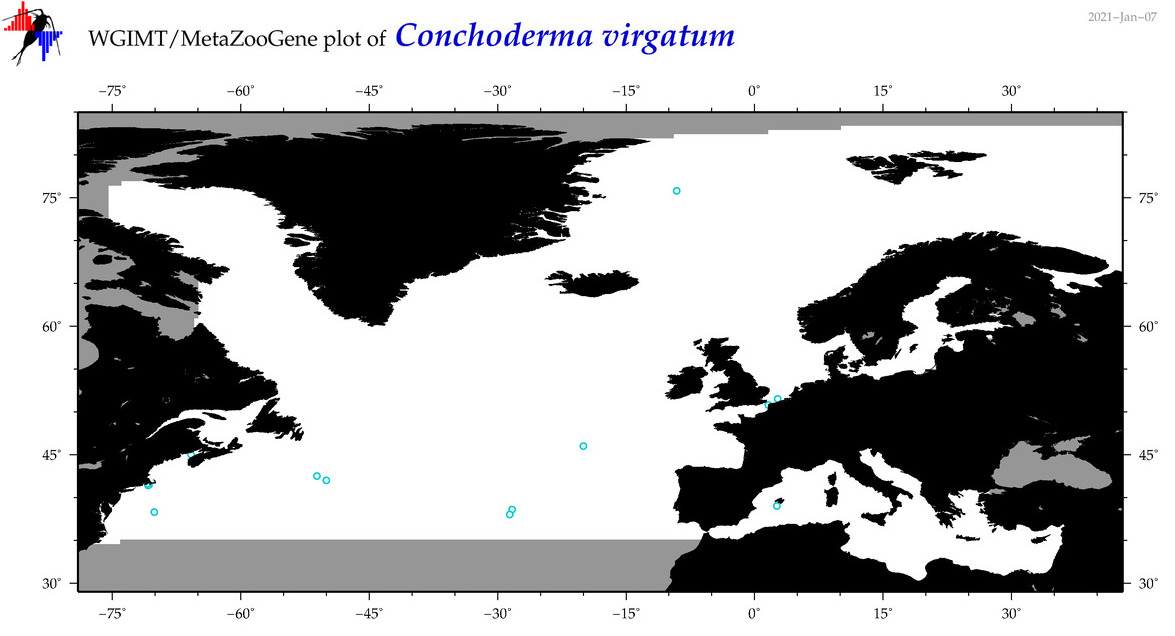 |
accepted
T4021713
R:1:0:0:0 |
| Coronula diadema |
Species
(14) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
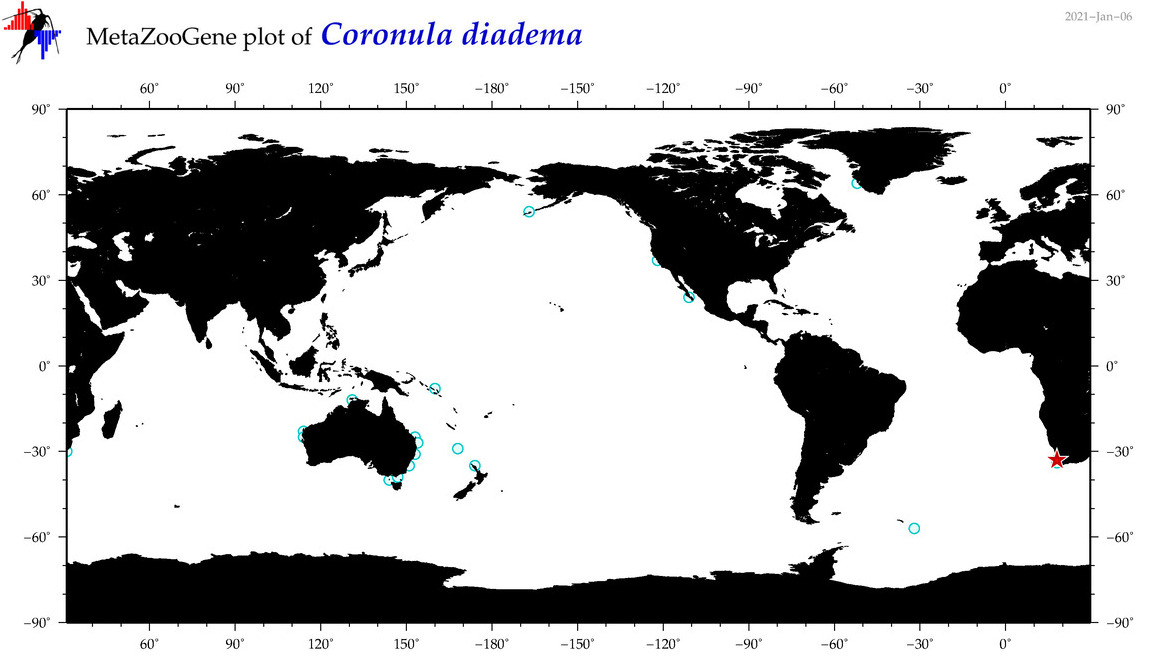 |
accepted
T4006480
R:1:0:0:0 |
| Dosima fascicularis |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 0
16S = 1
18S = 1
28S = 1
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
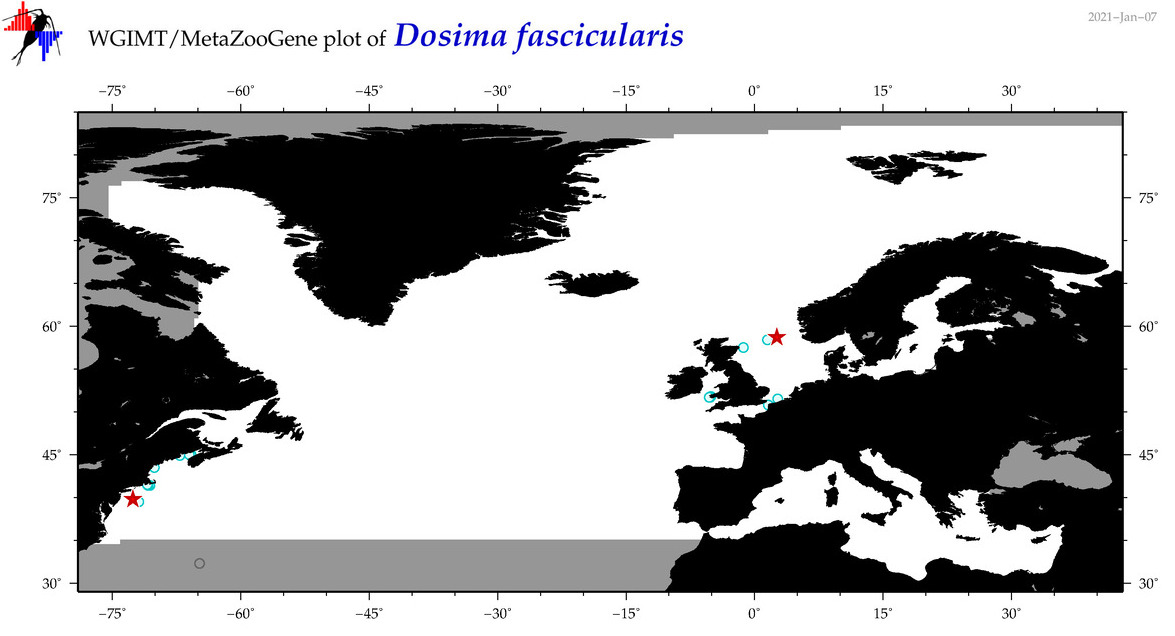 |
accepted
T4011294
R:1:0:0:0 |
| Fistulobalanus kondakovi |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 3
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
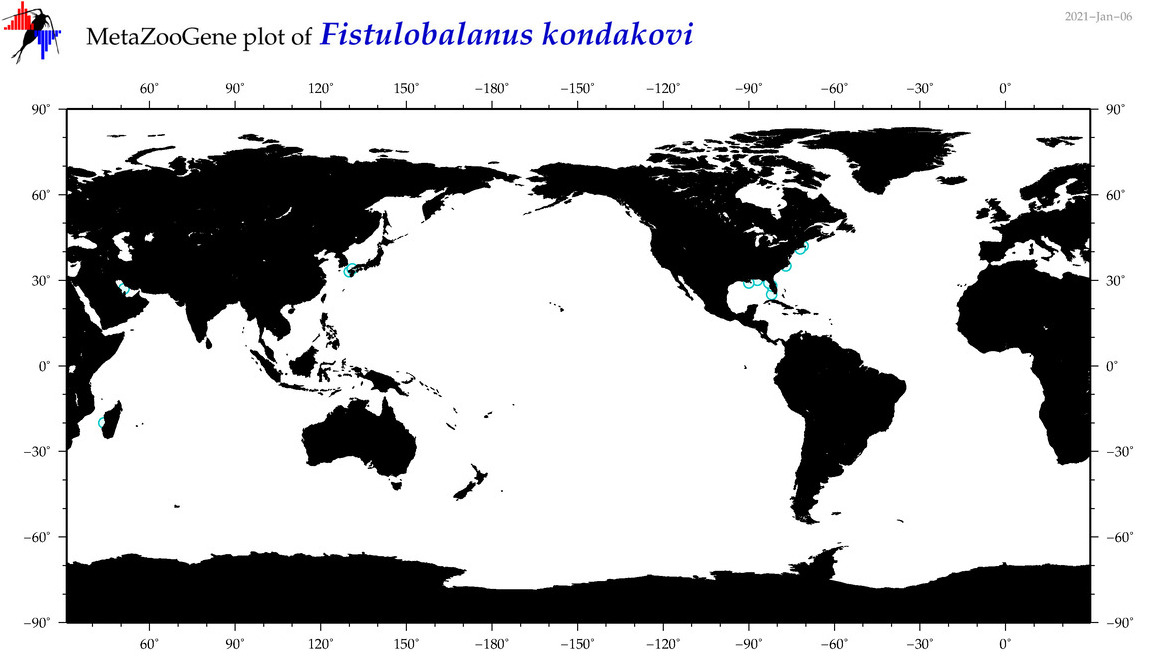 |
accepted
T4052299
R:1:0:0:0 |
| Lepas (Lepas) pectinata |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 118
12S = 0
16S = 12
18S = 2
28S = 1
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
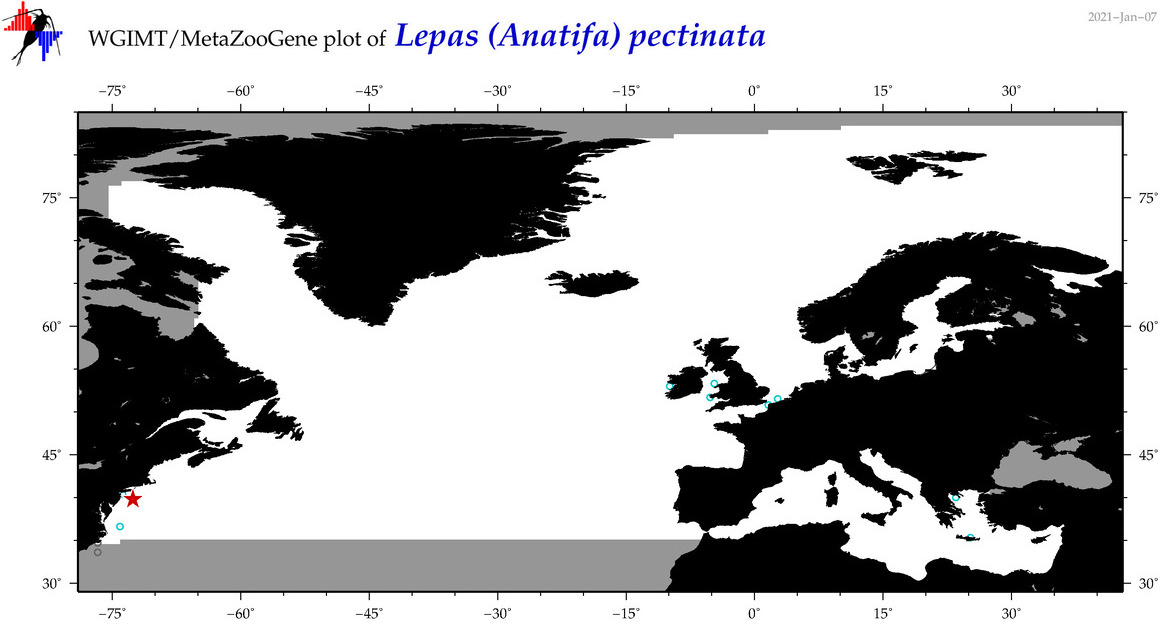 |
accepted
T4096249
R:1:0:0:0 |
| Loxothylacus panopaei |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 25
12S = 0
16S = 1
18S = 1
28S = 0
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
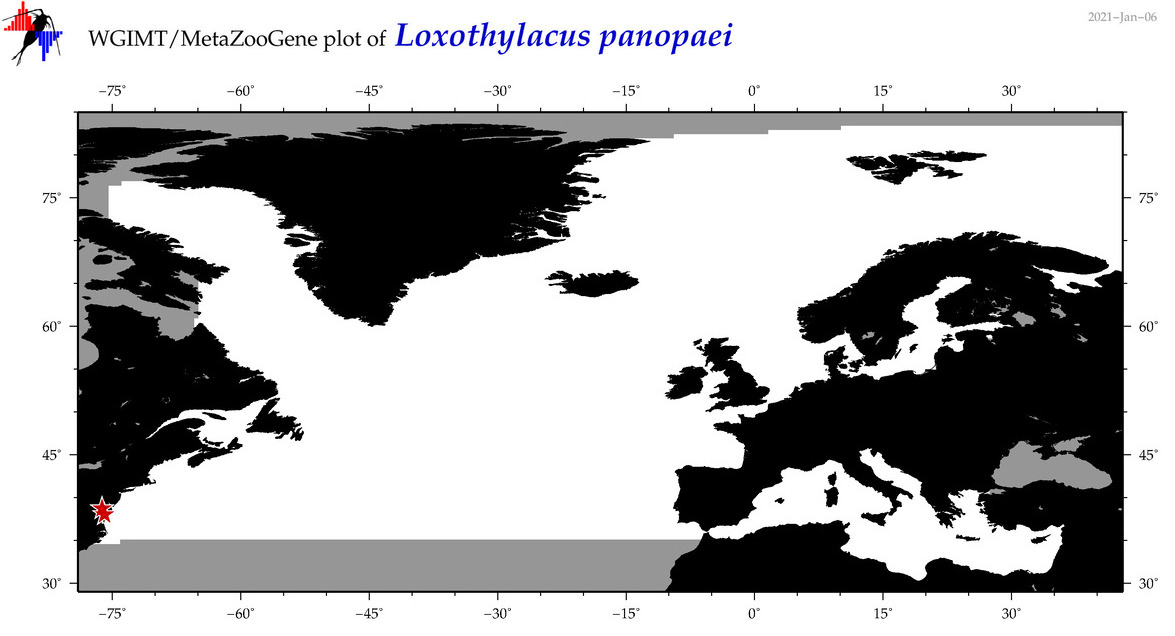 |
accepted
T4006749
R:1:0:0:0 |
| Octolasmis lowei |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
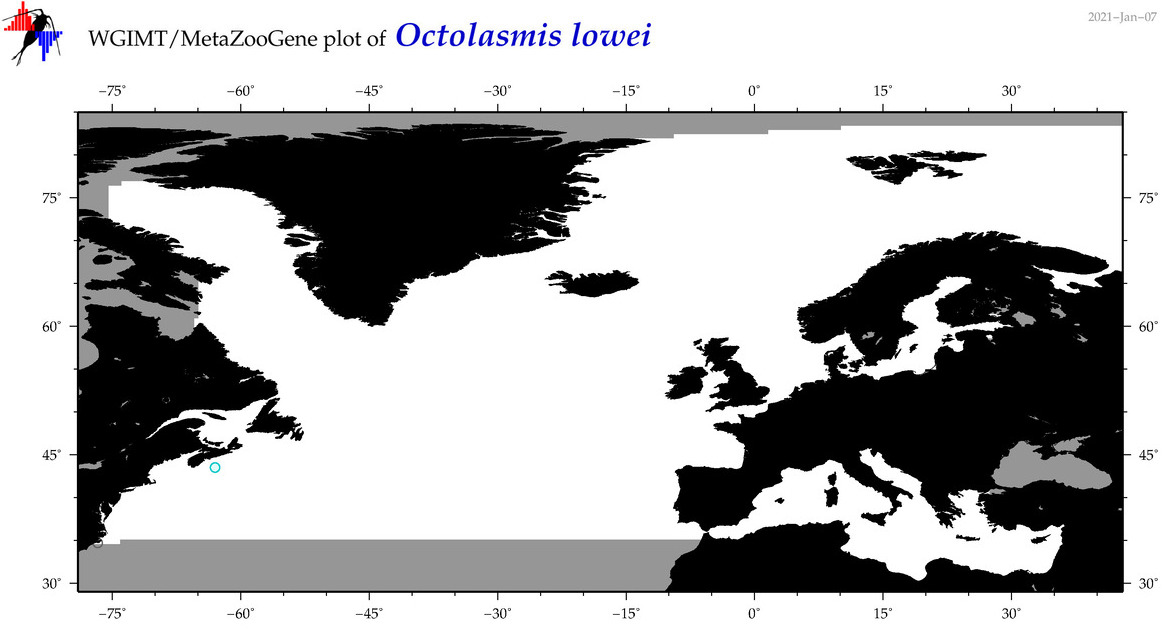 |
accepted
T4060813
R:1:0:0:0 |
| Poecilasma inaequilaterale |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
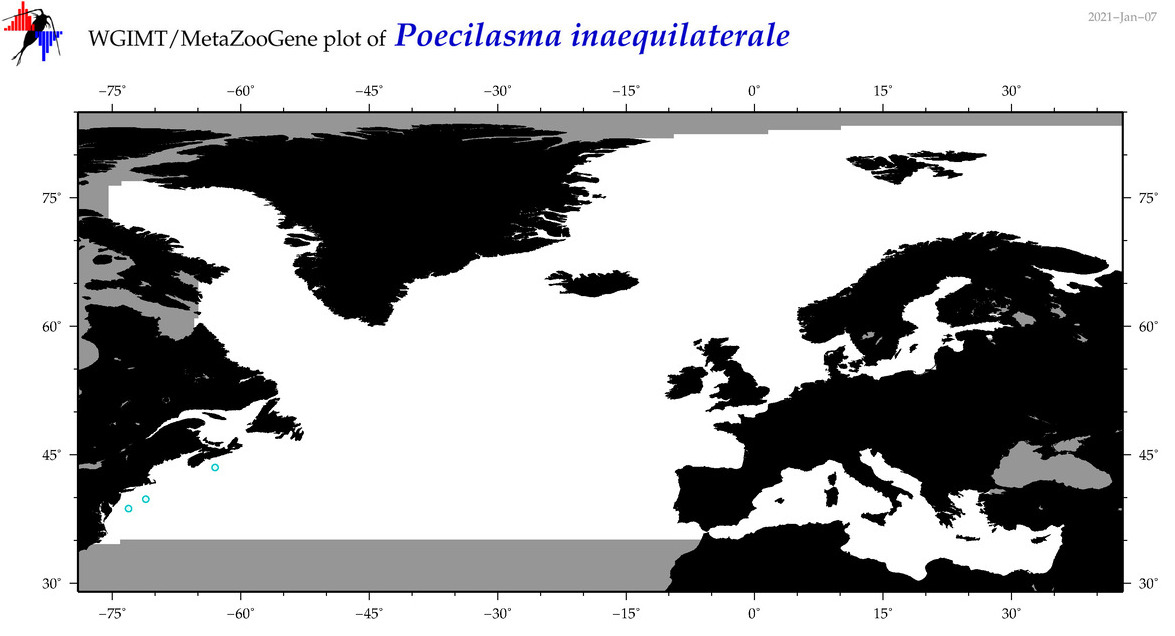 |
accepted
T4064601
R:1:0:0:0 |
| Semibalanus balanoides |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 943
12S = 10
16S = 17
18S = 7
28S = 2
|
COI = 55
12S = 0
16S = 0
18S = 0
28S = 0
|
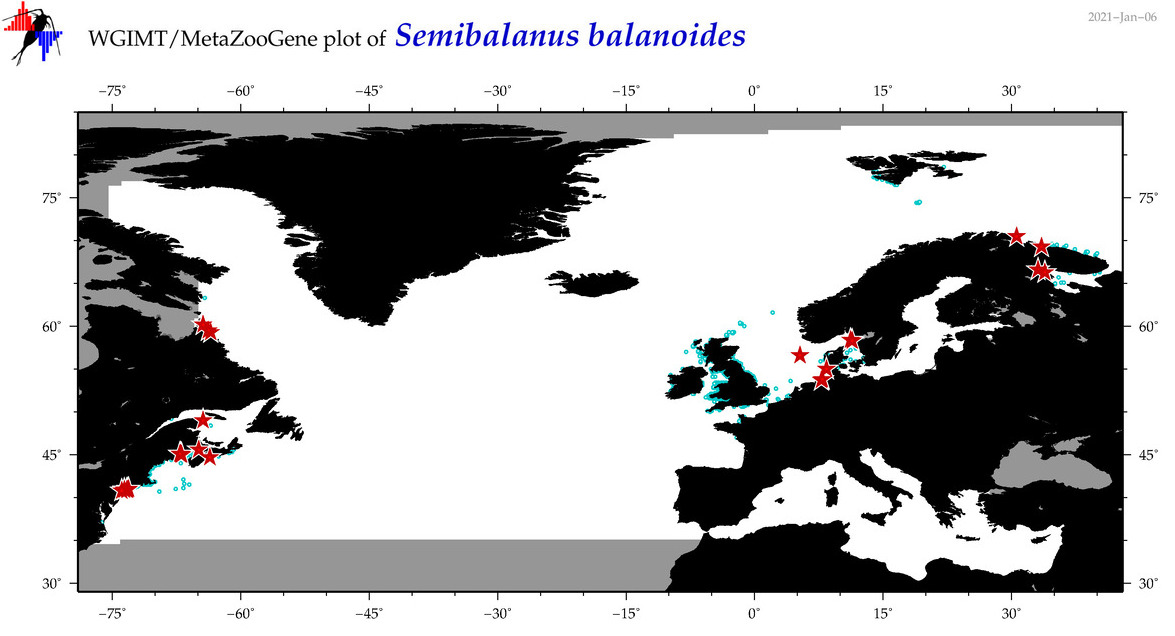 |
accepted
T4000517
R:1:0:0:0 |
| Sylon hippolytes |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
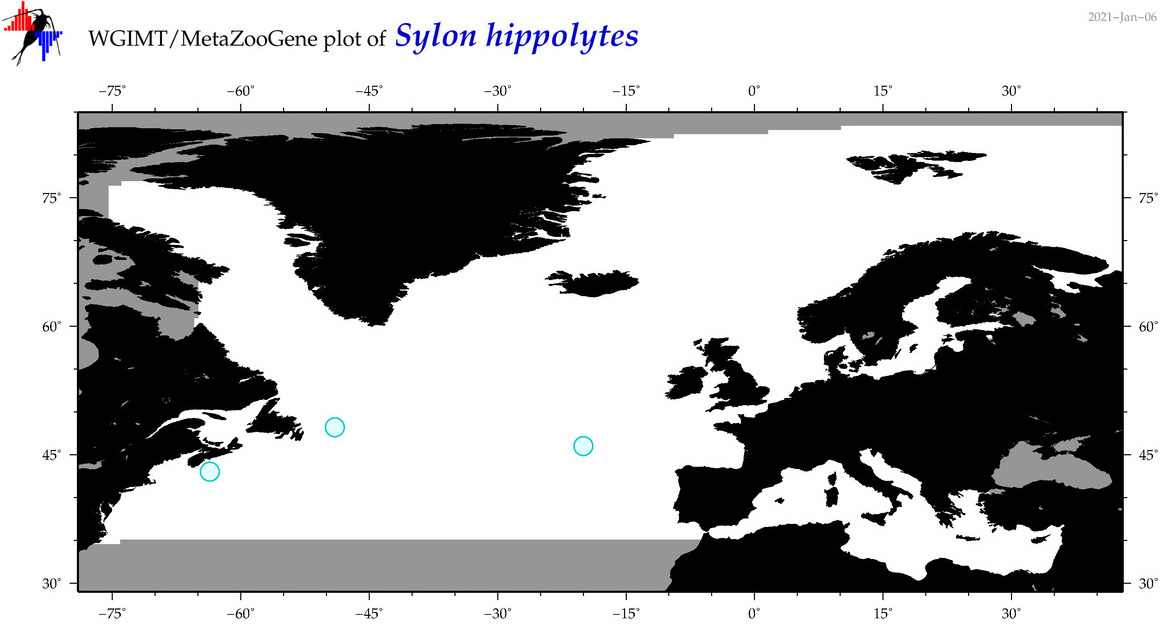 |
accepted
T4032277
R:1:0:0:0 |