Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-26 |
| Bathycuma brevirostre |
Species
(1) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
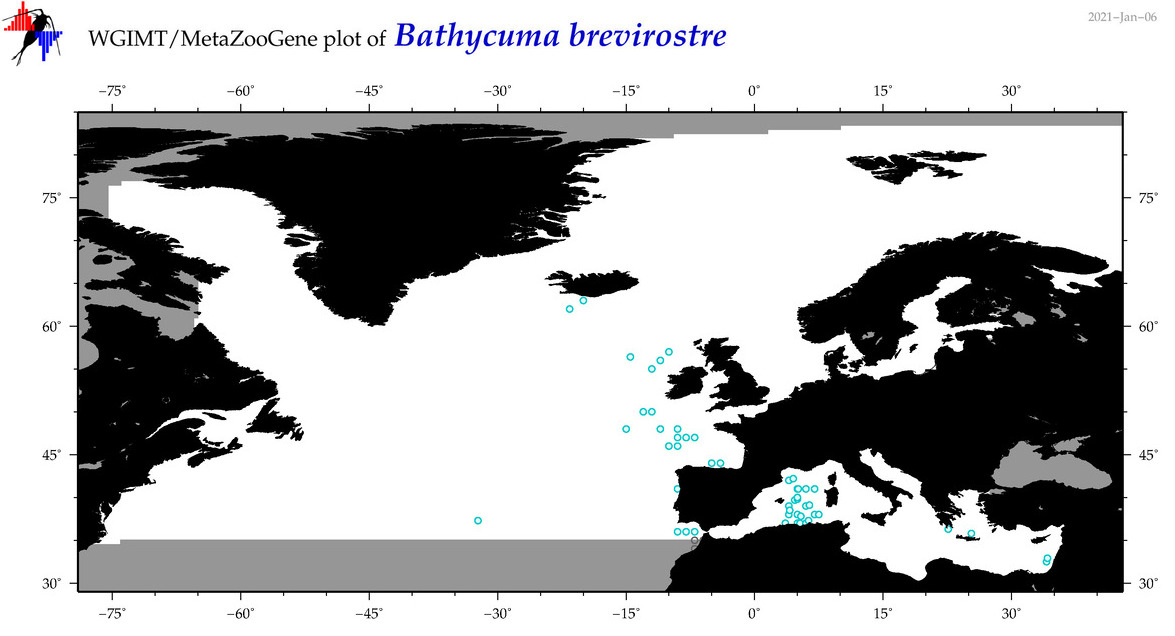 |
accepted
T4045194
R:1:0:0:0 |
| Campylaspis affinis |
Species
(2) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
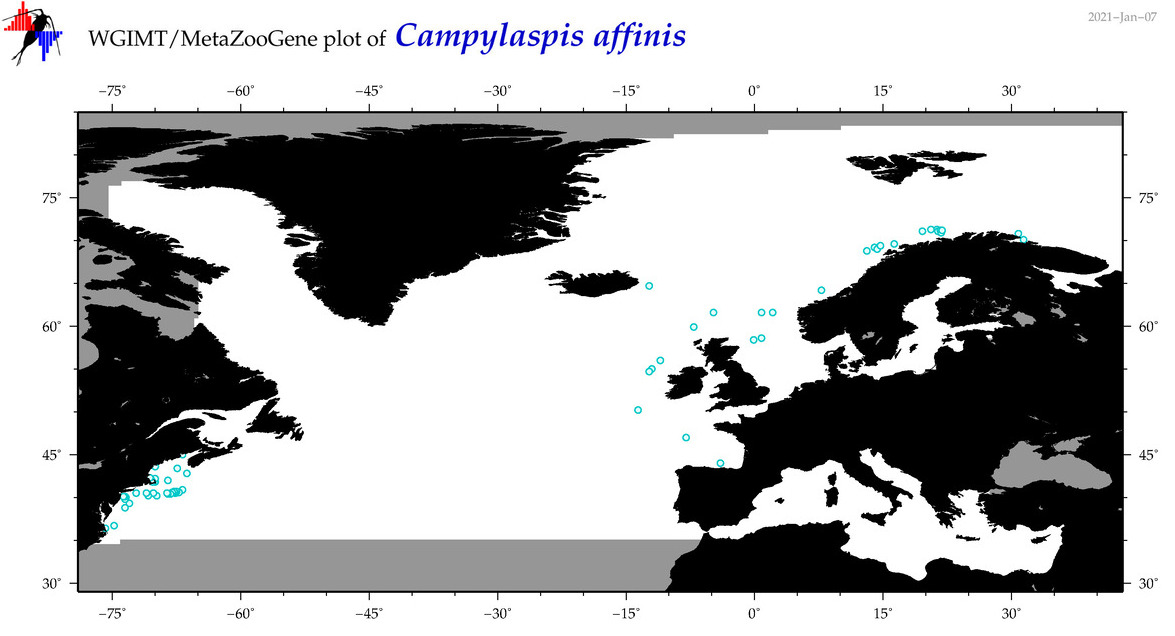 |
accepted
T4046523
R:1:0:0:0 |
| Campylaspis alba |
Species
(3) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
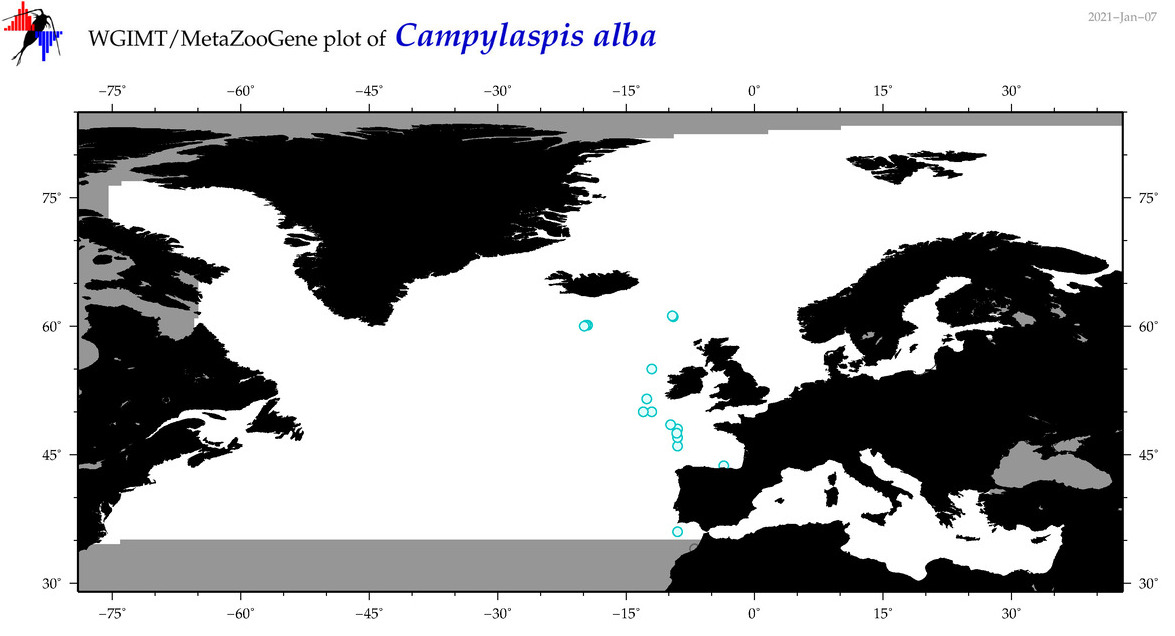 |
accepted
T4046525
R:1:0:0:0 |
| Campylaspis globosa |
Species
(4) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4046556
R:1:0:0:0 |
| Campylaspis intermedia |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
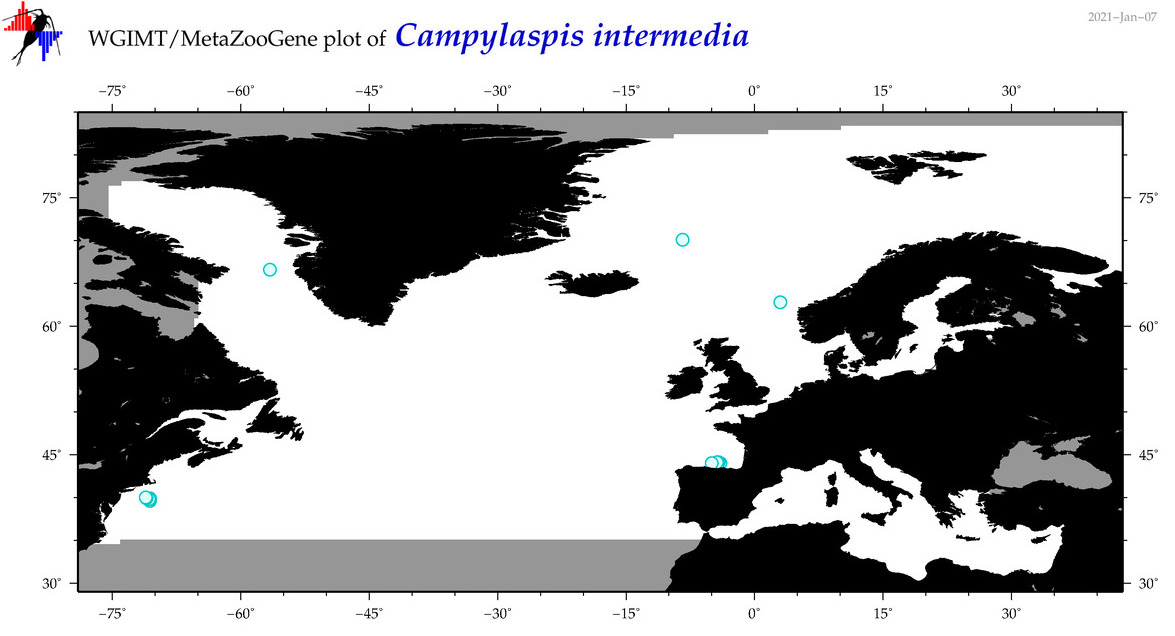 |
accepted
T4046568
R:1:0:0:0 |
| Campylaspis mansa |
Species
(6) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4046584
R:1:0:0:0 |
| Campylaspis rubicunda |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 7
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
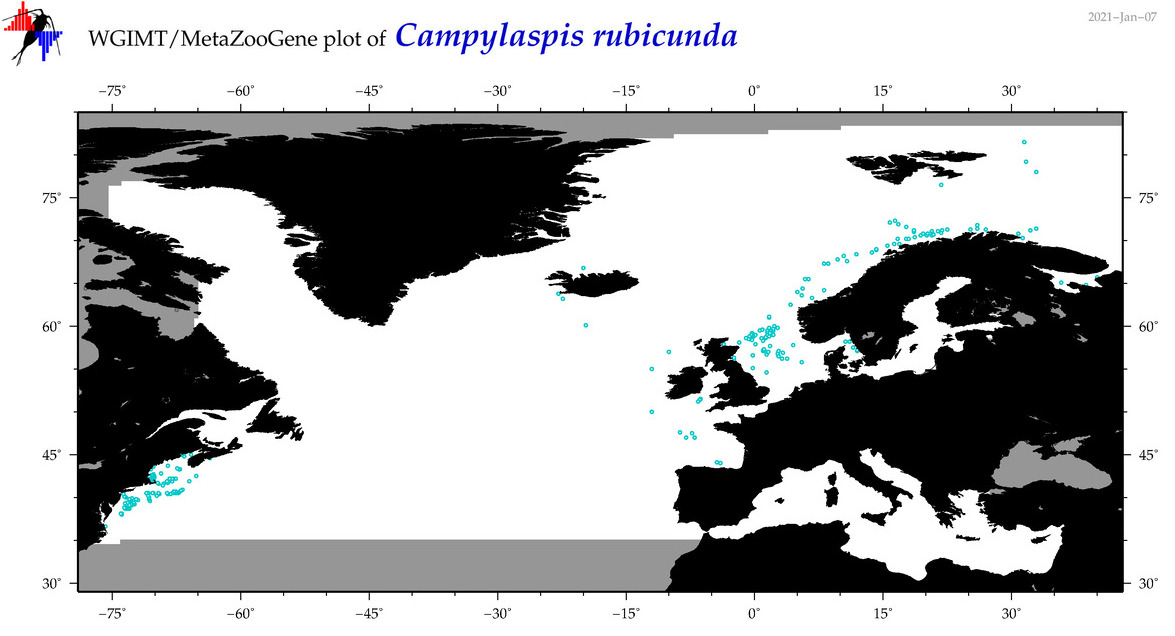 |
accepted
T4046615
R:1:0:0:0 |
| Campylaspis valleculata |
Species
(8) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
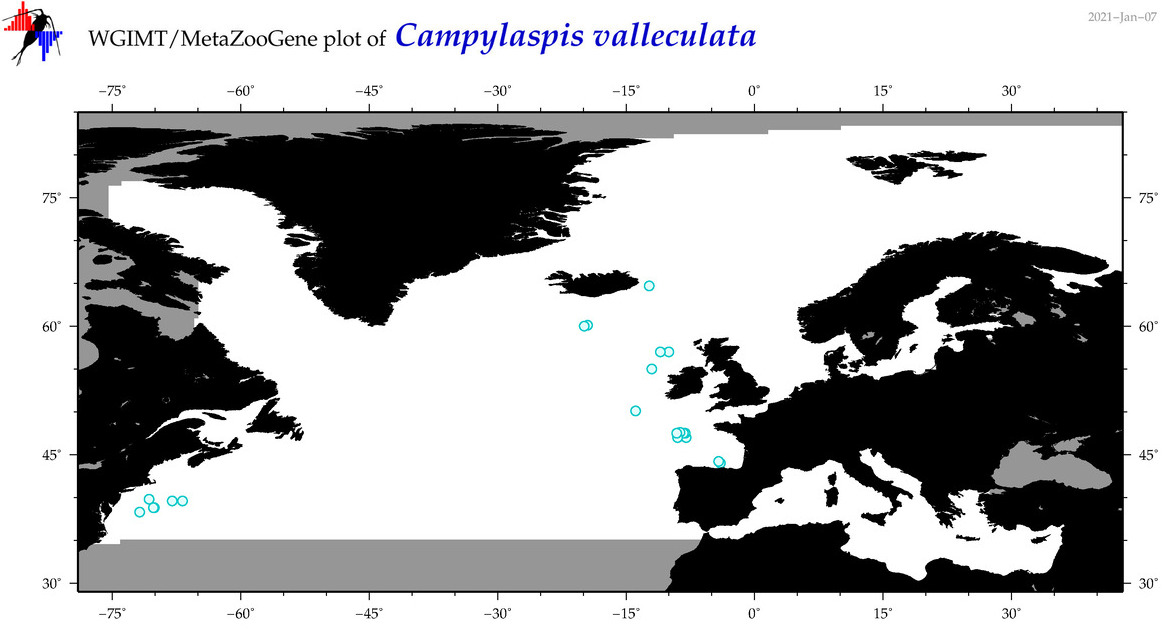 |
accepted
T4046642
R:1:0:0:0 |
| Campylaspis verrucosa |
Species
(9) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
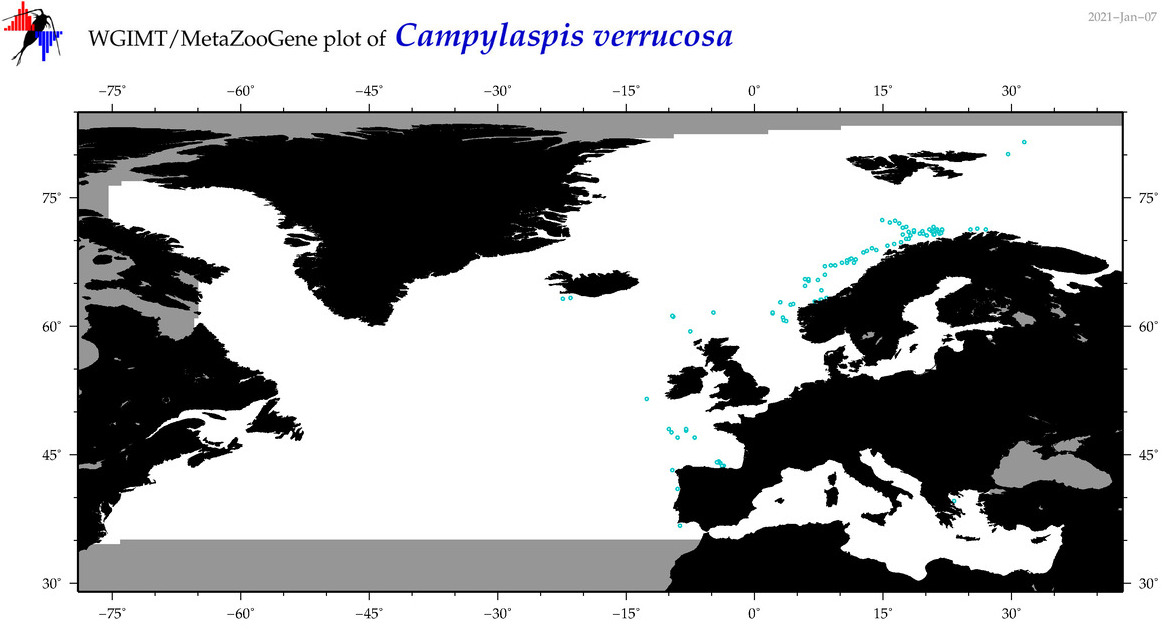 |
accepted
T4046643
R:1:0:0:0 |
| Cimmerius reticulatus |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 2
18S = 0
28S = 0
|
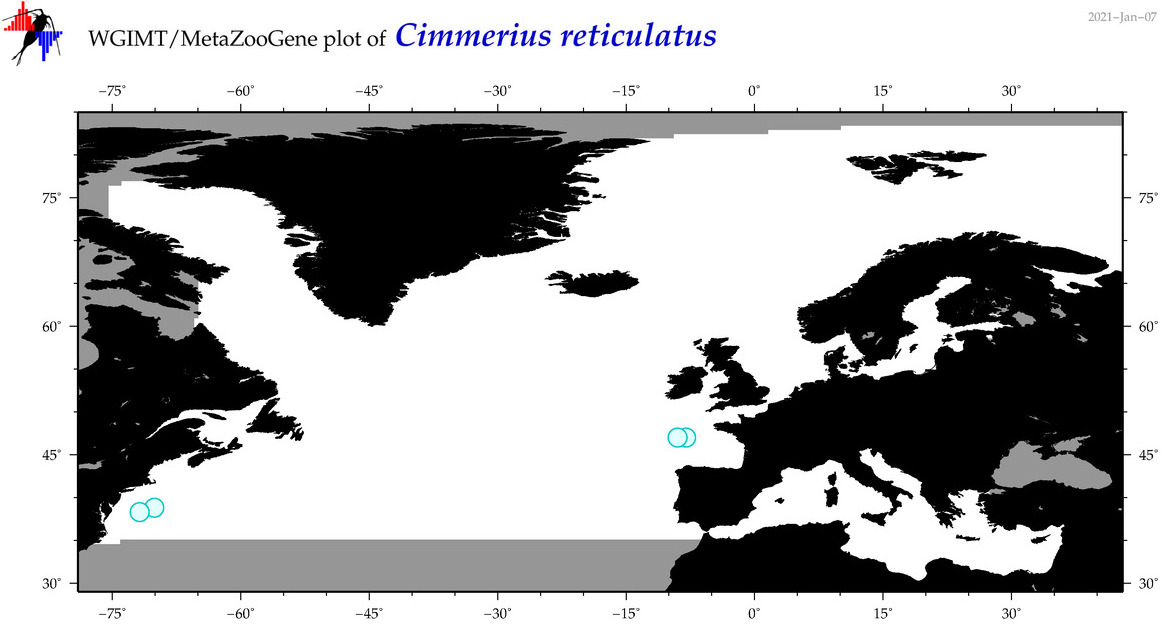 |
accepted
T4077641
R:1:0:0:0 |
| Cumella (Cumella) decipiens |
Species
(11) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
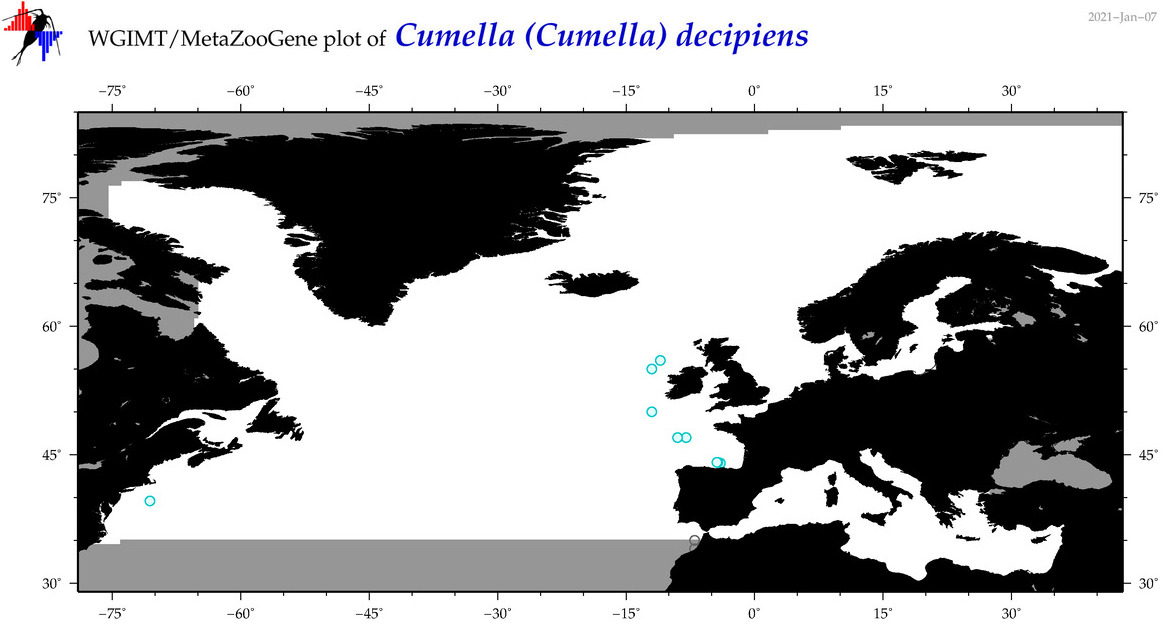 |
accepted
T4091474
R:1:0:0:0 |
| Cumellopsis bicostata |
Species
(12) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
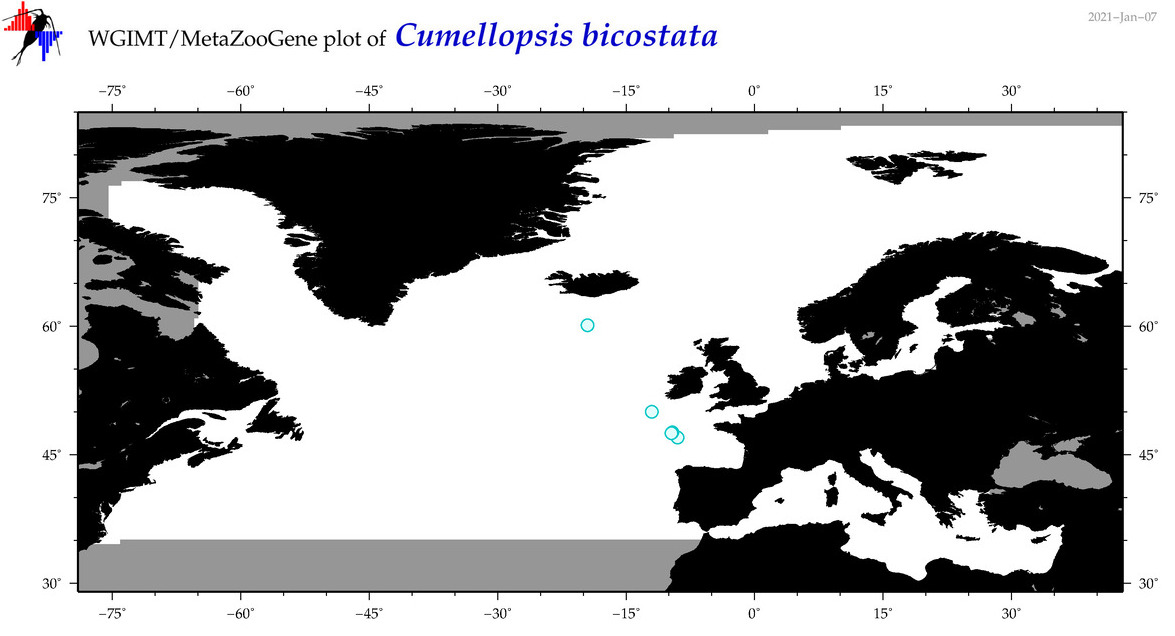 |
accepted
T4048823
R:1:0:0:0 |
| Cumellopsis helgae |
Species
(13) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
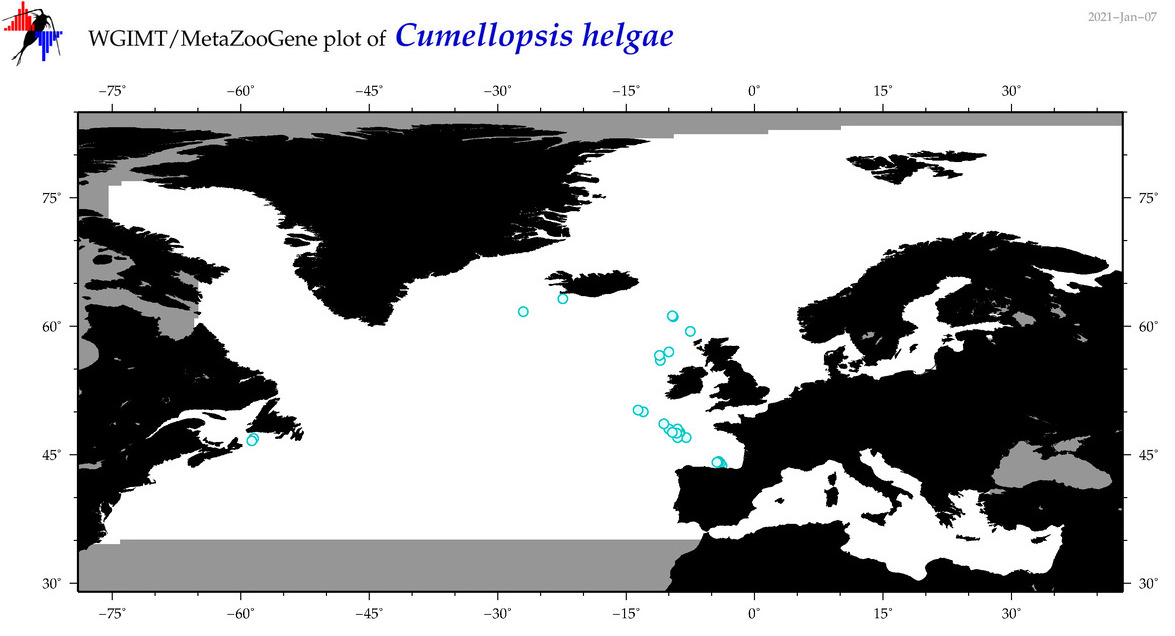 |
accepted
T4048825
R:1:0:0:0 |
| Cumellopsis puritani |
Species
(14) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
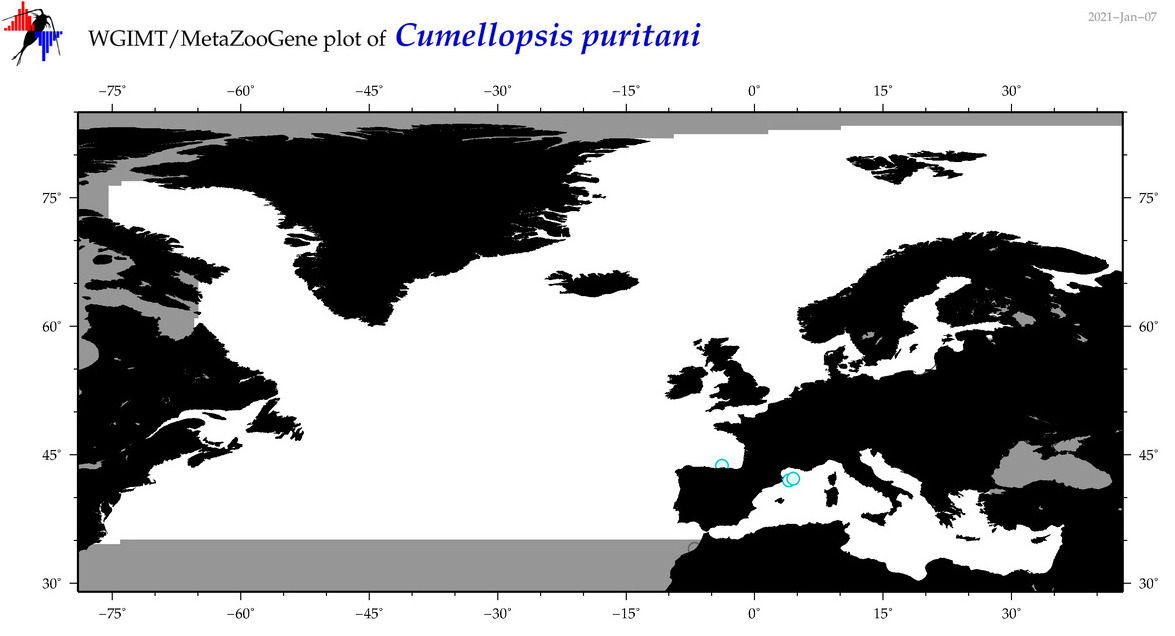 |
accepted
T4048827
R:1:0:0:0 |
| Cyclaspis longicaudata |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
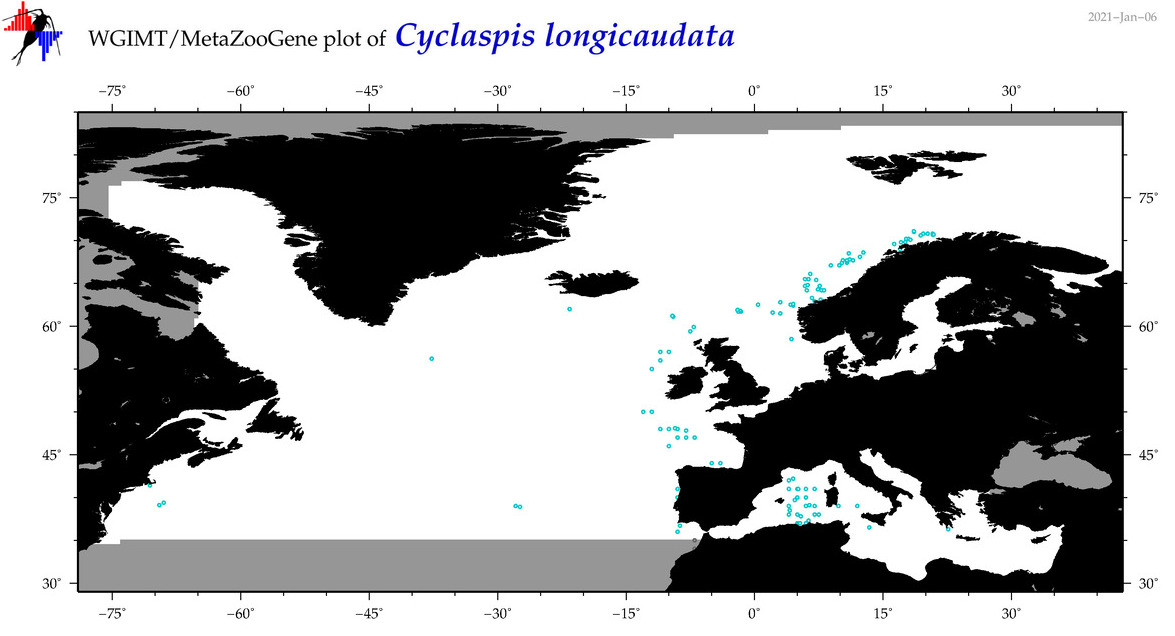 |
accepted
T4048905
R:1:0:0:0 |
| Diastylis goodsiri |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 16
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4016714
R:1:1:0:0 |
| Diastylis polaris |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 6
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 6
18S = 0
28S = 0
|
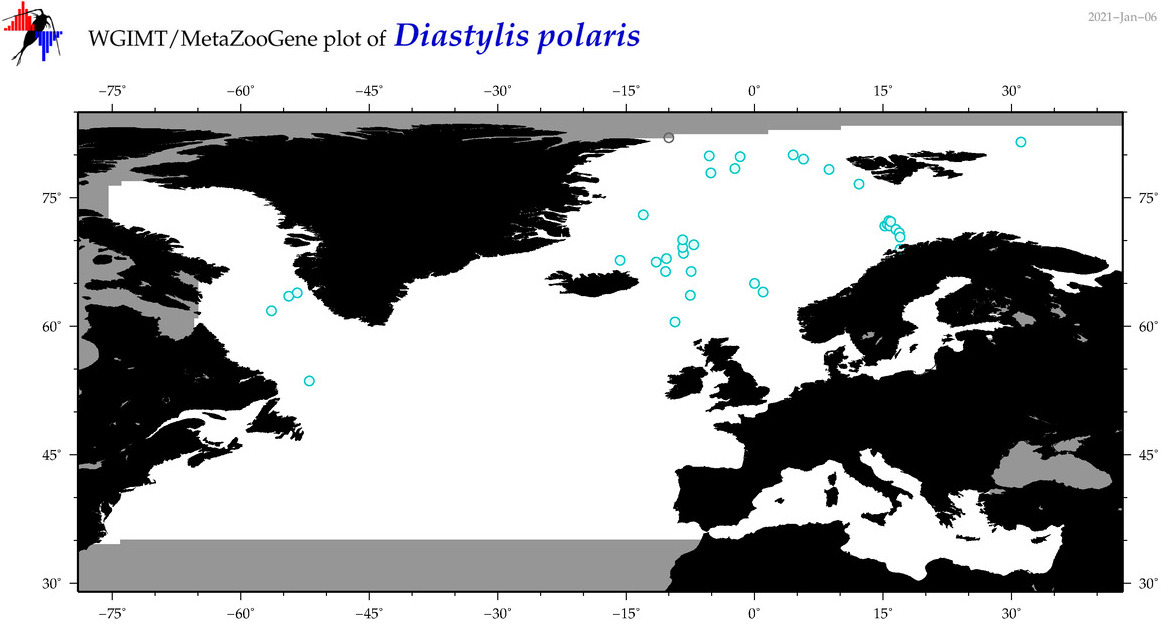 |
accepted
T4049615
R:1:1:0:0 |
| Diastylis rathkei |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4011179
R:1:1:0:0 |
| Diastylis scorpioides |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
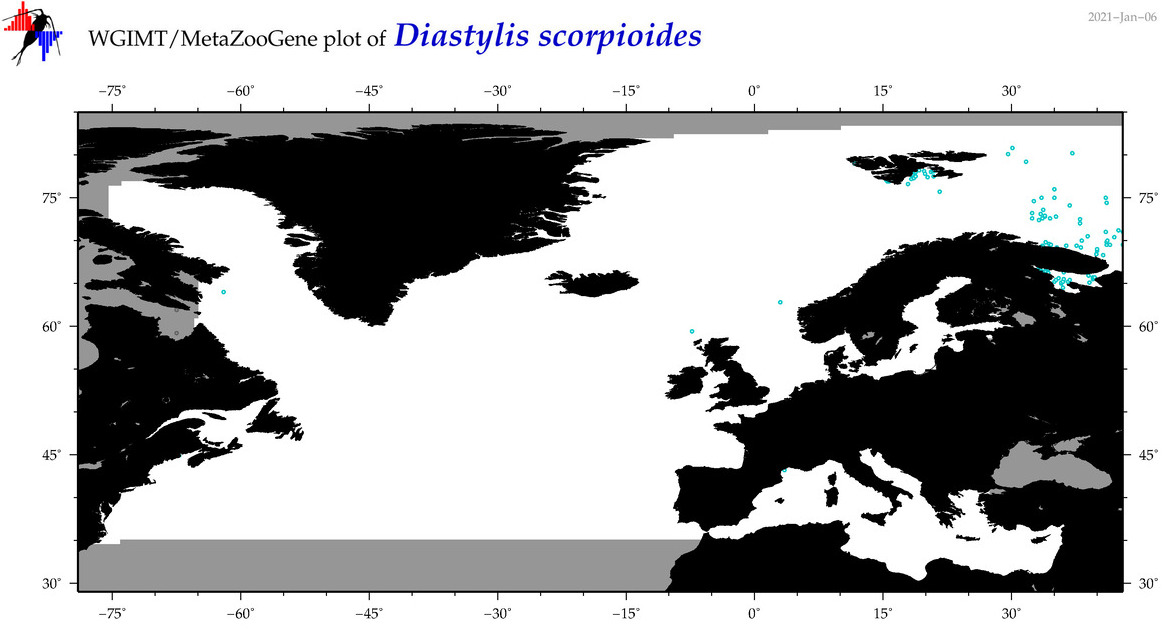 |
accepted
T4049622
R:1:1:0:0 |
| Diastylis spinulosa |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
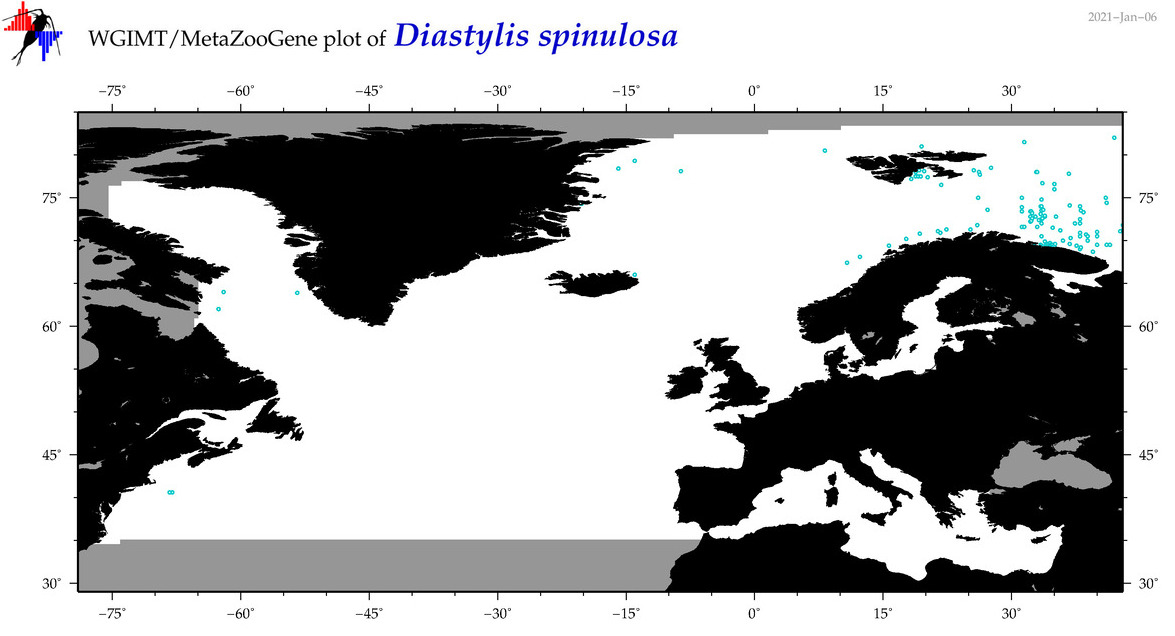 |
accepted
T4049625
R:1:1:0:0 |
| Eudorella emarginata |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 162
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
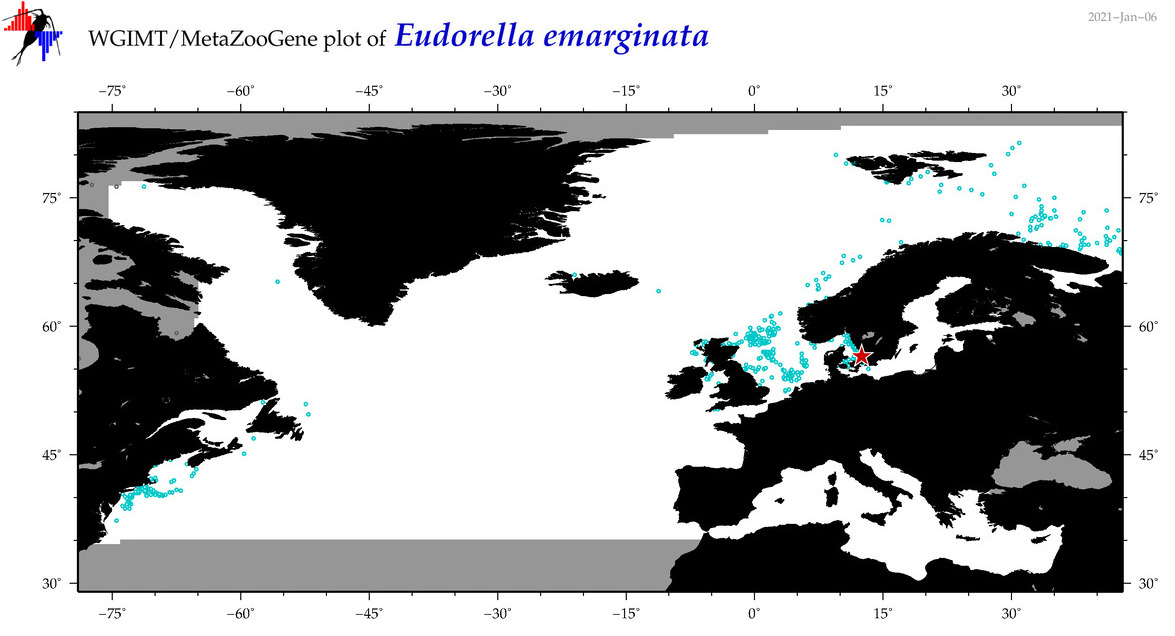 |
accepted
T4016749
R:1:1:0:0 |
| Eudorella hispida |
Species
(22) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
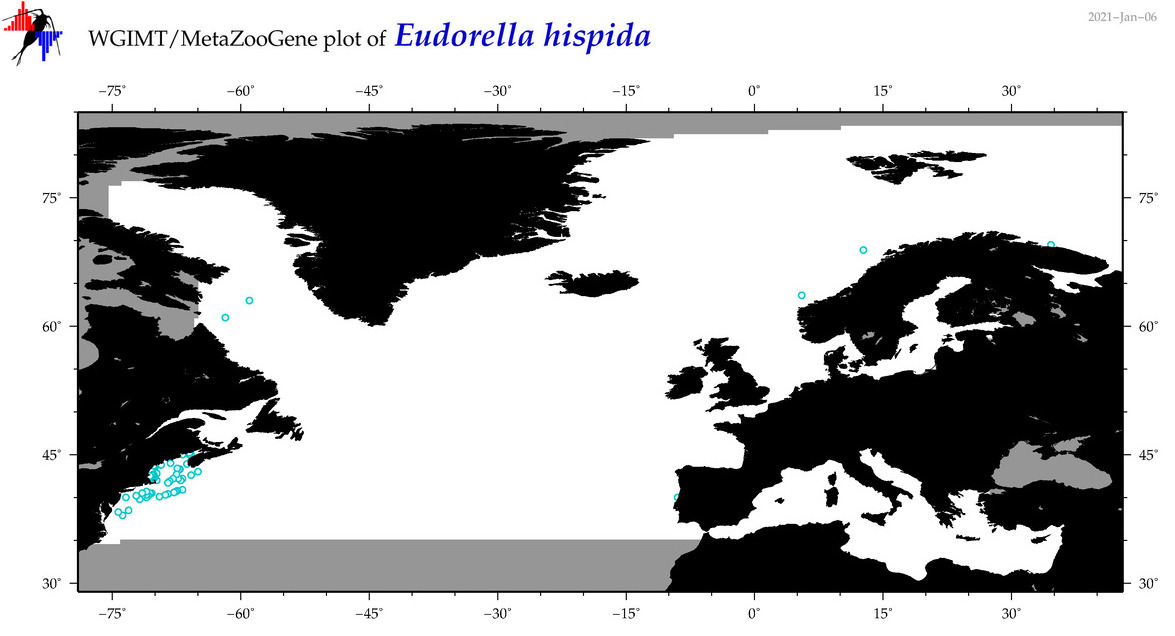 |
accepted
T4051392
R:1:1:0:0 |
| Hemilamprops cristatus |
Species
(23) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
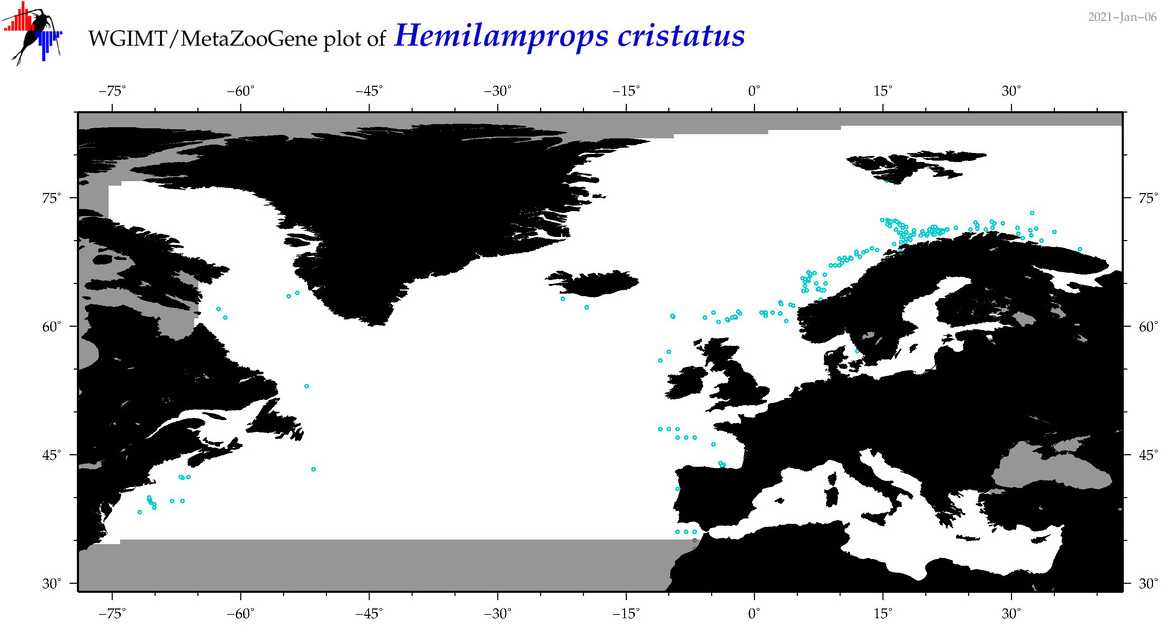 |
accepted
T4053844
R:1:0:0:0 |
| Hemilamprops diversus |
Species
(24) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
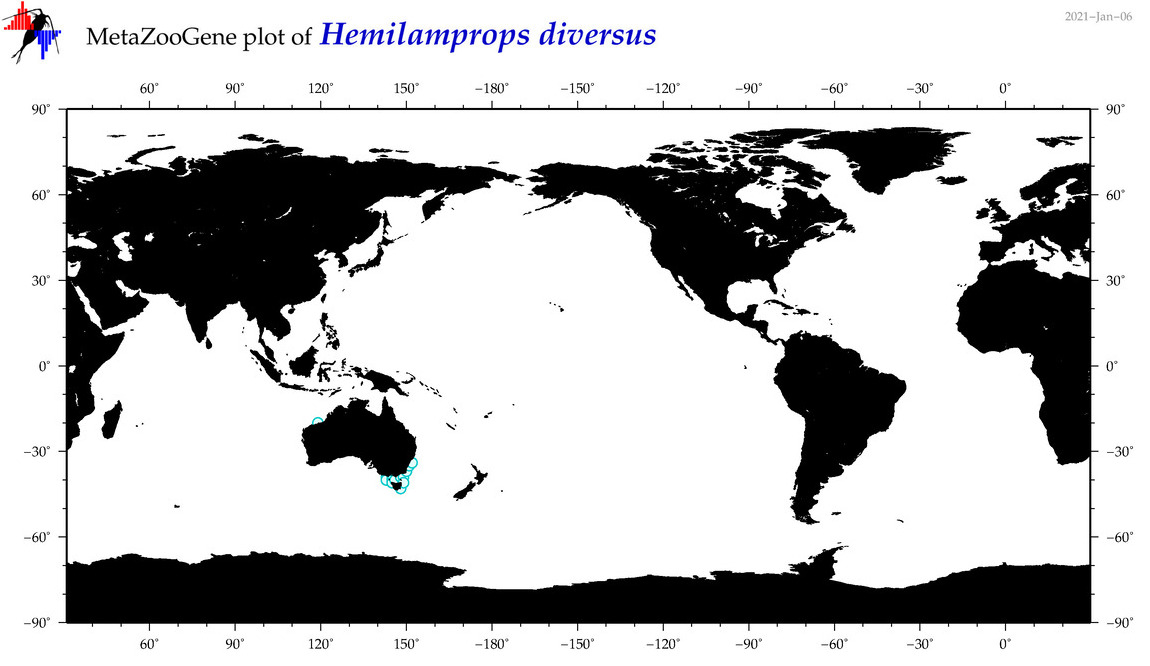 |
accepted
T4053845
R:1:0:0:0 |
| Hemilamprops pterini |
Species
(25) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 2
18S = 0
28S = 0
|
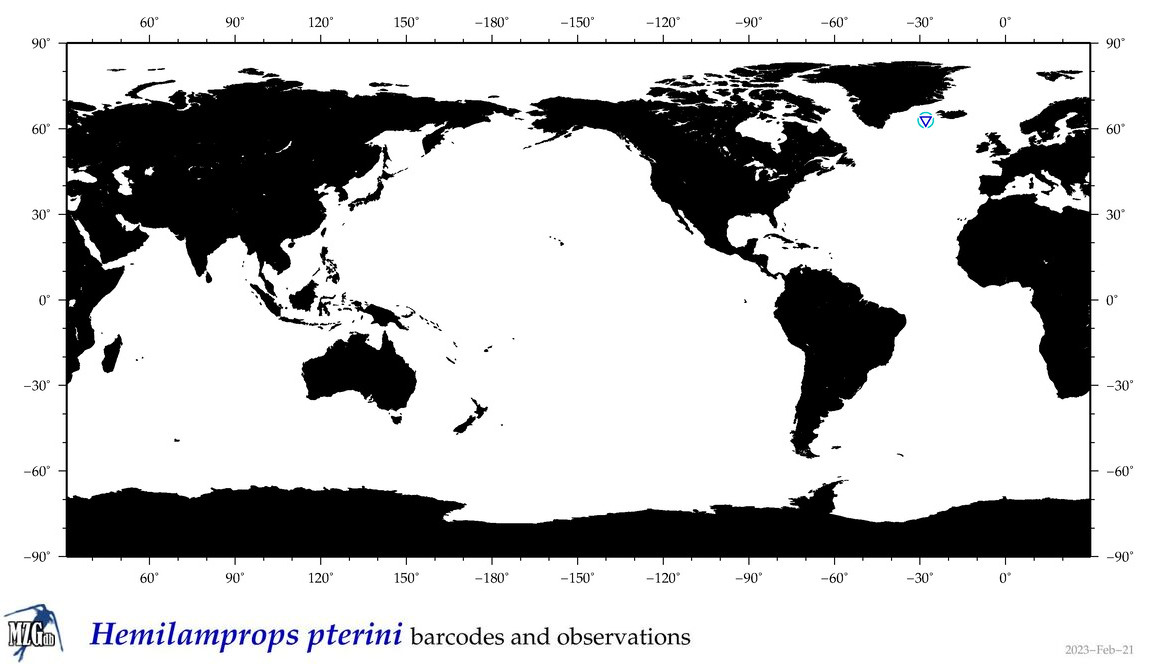 |
accepted
T4003095
R:1:0:0:0 |
| Hemilamprops uniplicatus |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
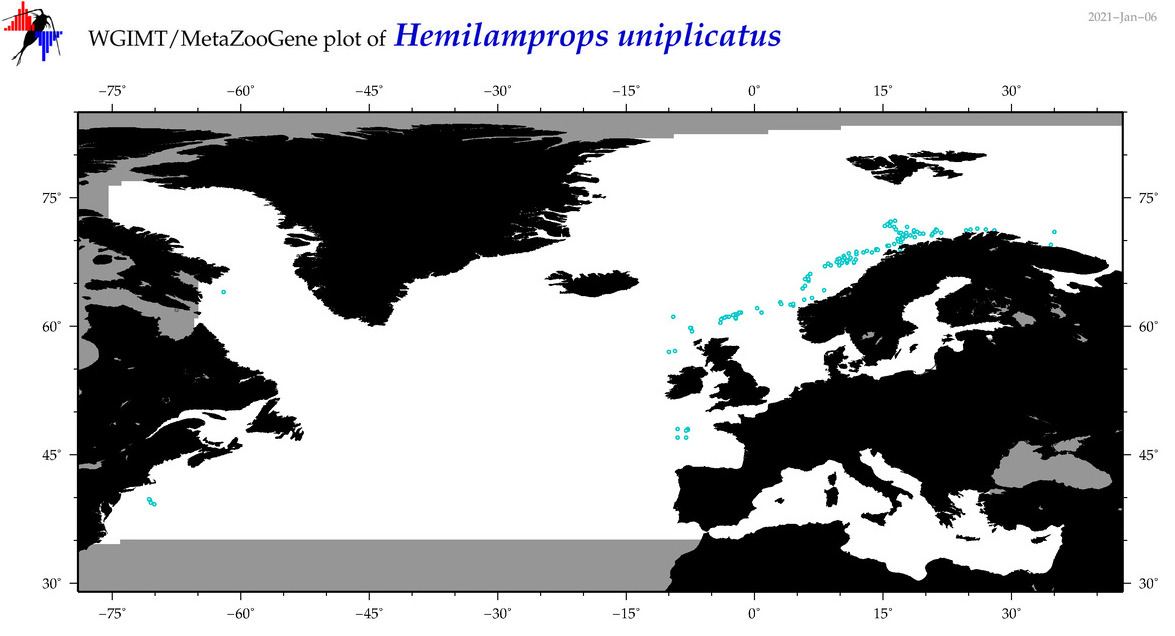 |
accepted
T4053854
R:1:0:0:0 |
| Leptostylis ampullacea |
Species
(27) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
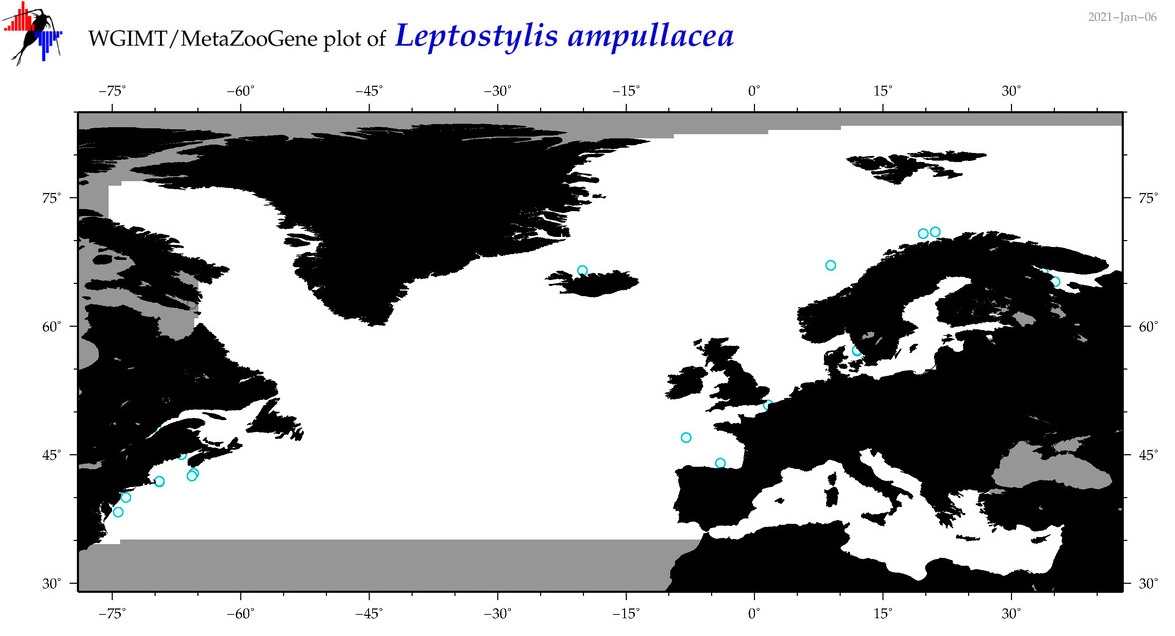 |
accepted
T4056228
R:1:1:0:0 |
| Leptostylis longimana |
Species
(28) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
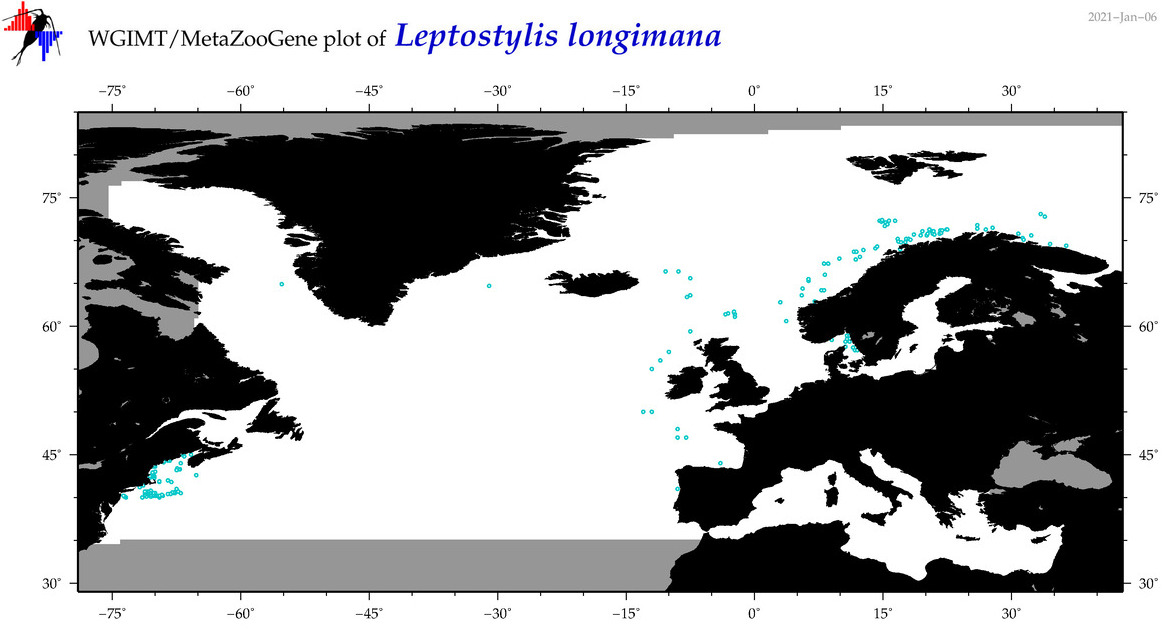 |
accepted
T4056245
R:1:1:0:0 |
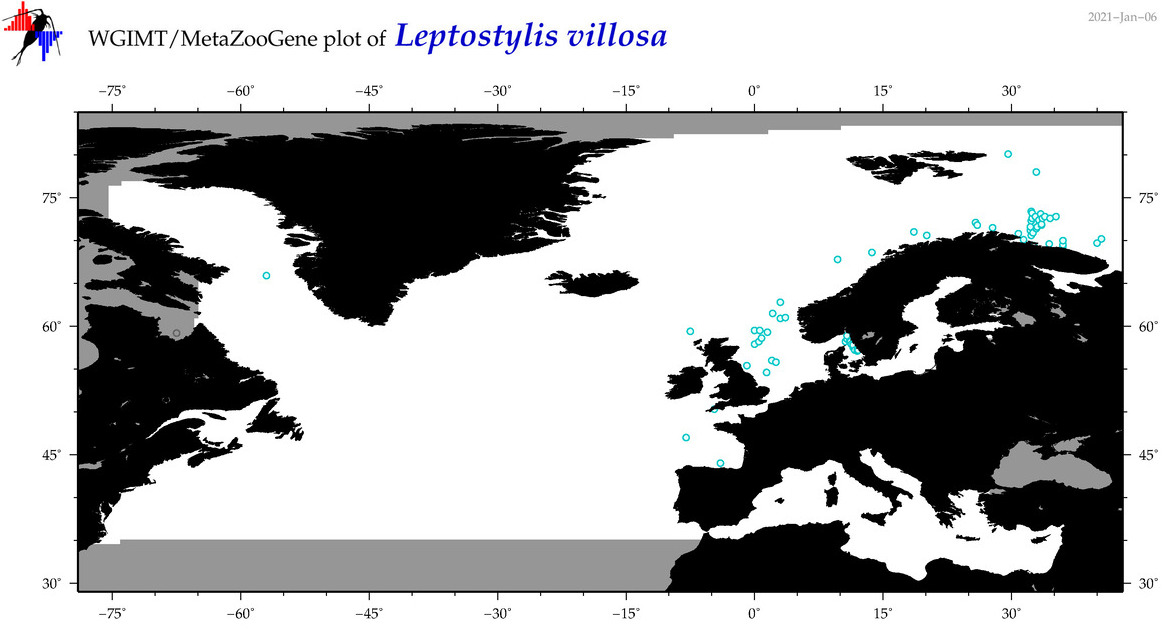
| Leptostylis villosa |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4056253
R:1:1:0:0 |
| Leucon (Alytoleucon) pallidus |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 7
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 5
18S = 0
28S = 0
|
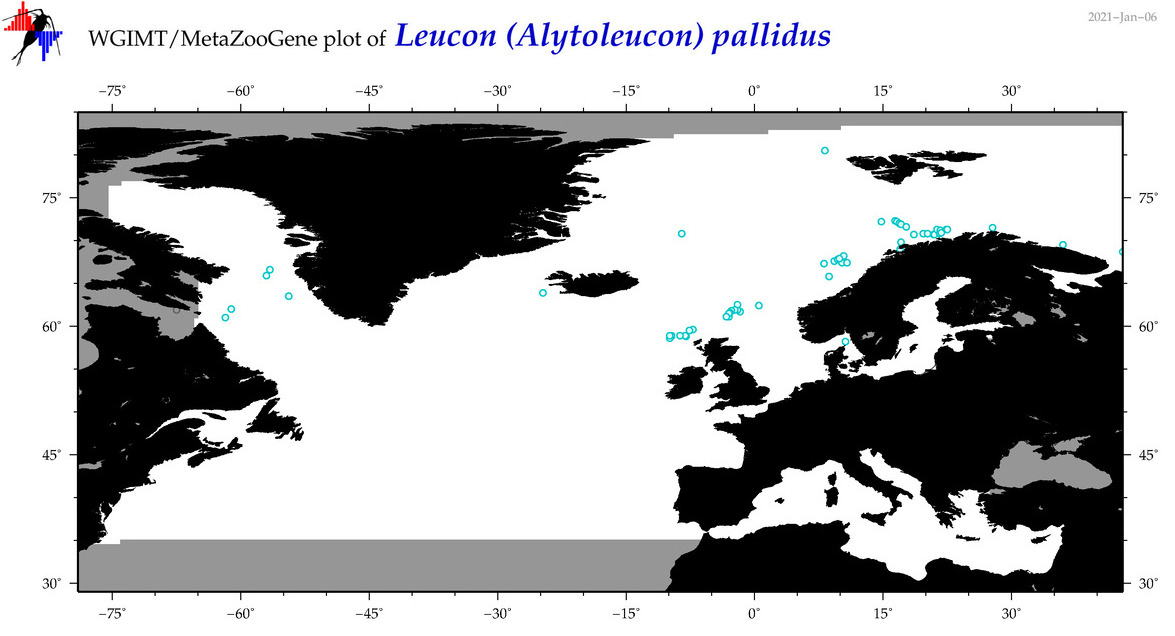 |
accepted
T4091486
R:1:1:0:0 |
| Leucon (Crymoleucon) tener |
Species
(31) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
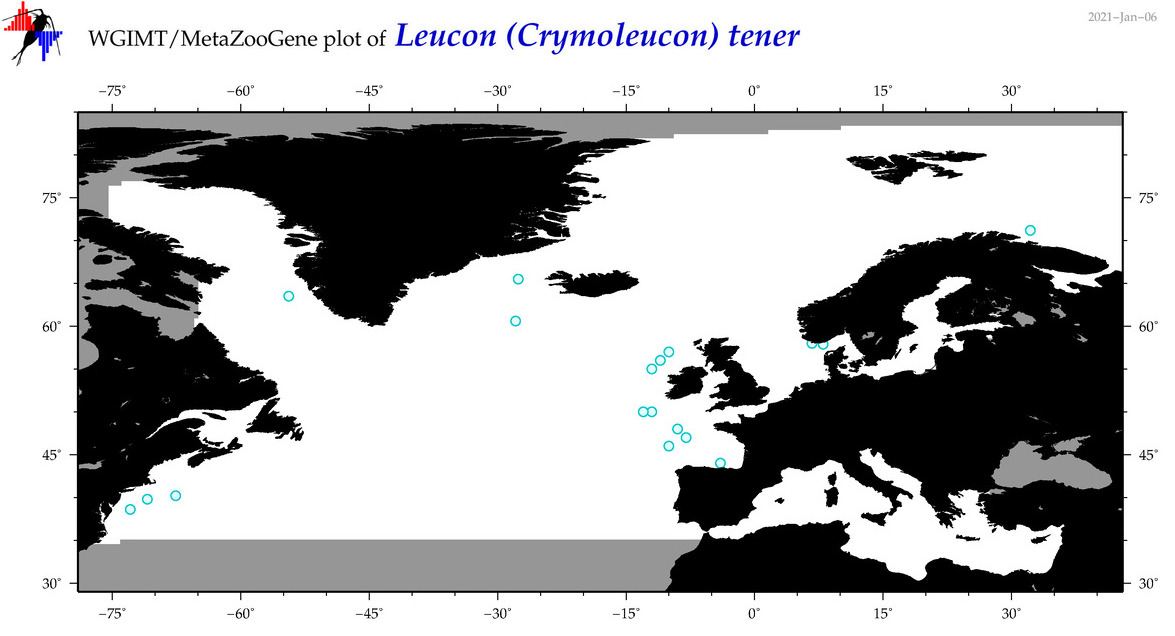 |
accepted
T4091488
R:1:1:0:0 |
| Leucon (Epileucon) spiniventris |
Species
(32) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
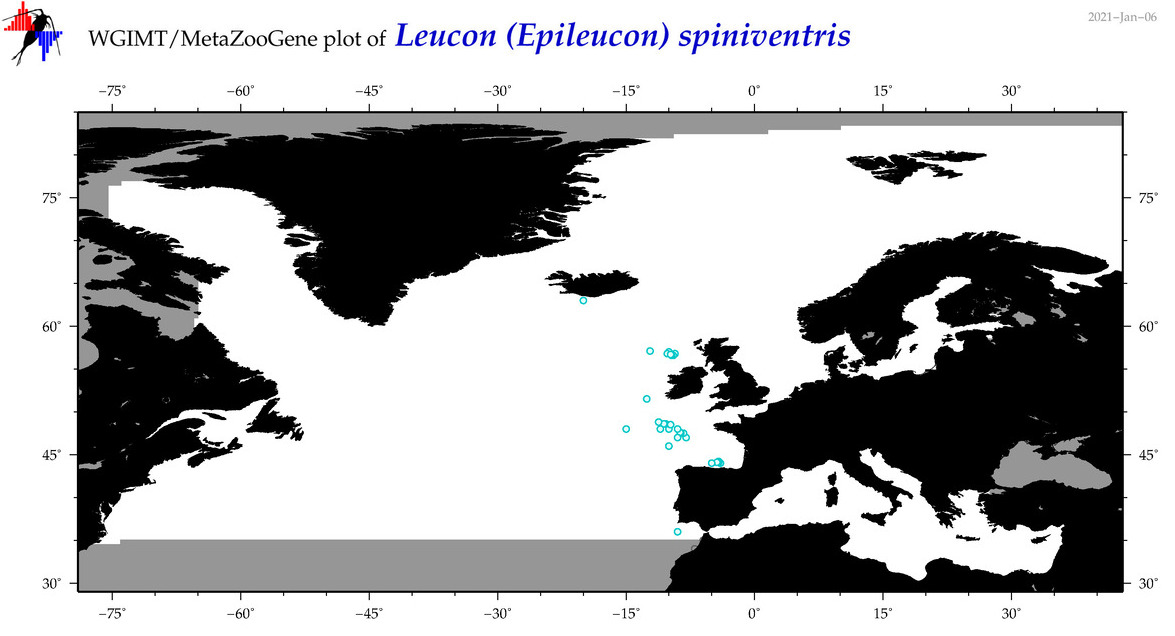 |
accepted
T4094855
R:1:1:0:0 |
| Leucon (Leucon) fulvus |
Species
(33) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4091492
R:1:1:0:0 |
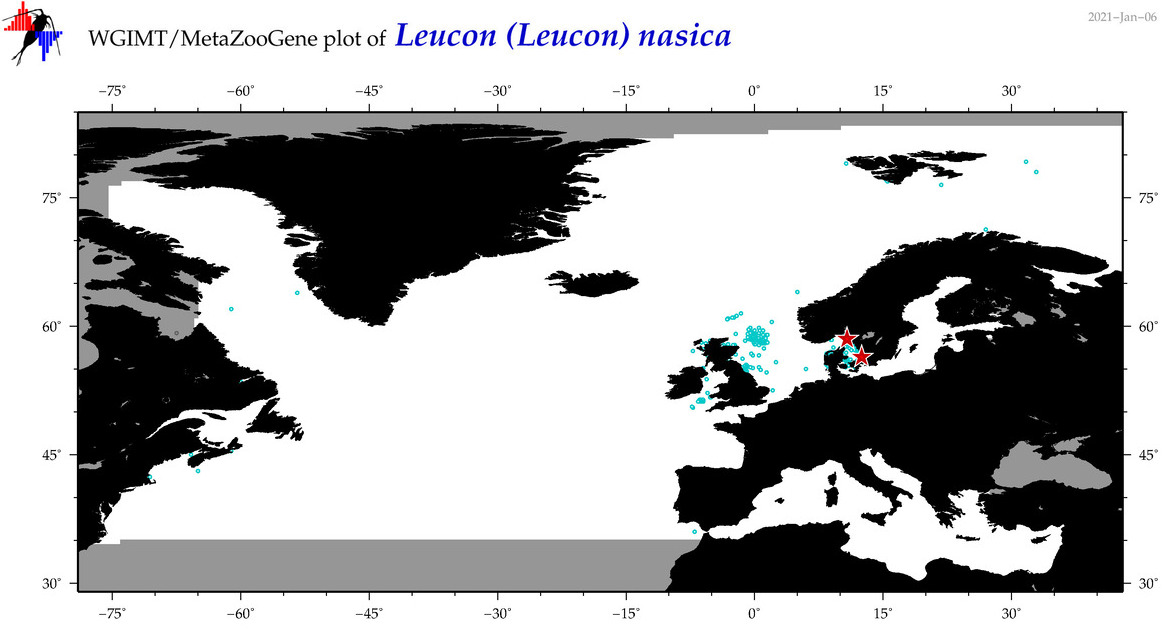
| Leucon (Leucon) nasica |
Species
(34) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4091494
R:1:1:0:0 |
| Leucon (Leucon) profundus |
Species
(35) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
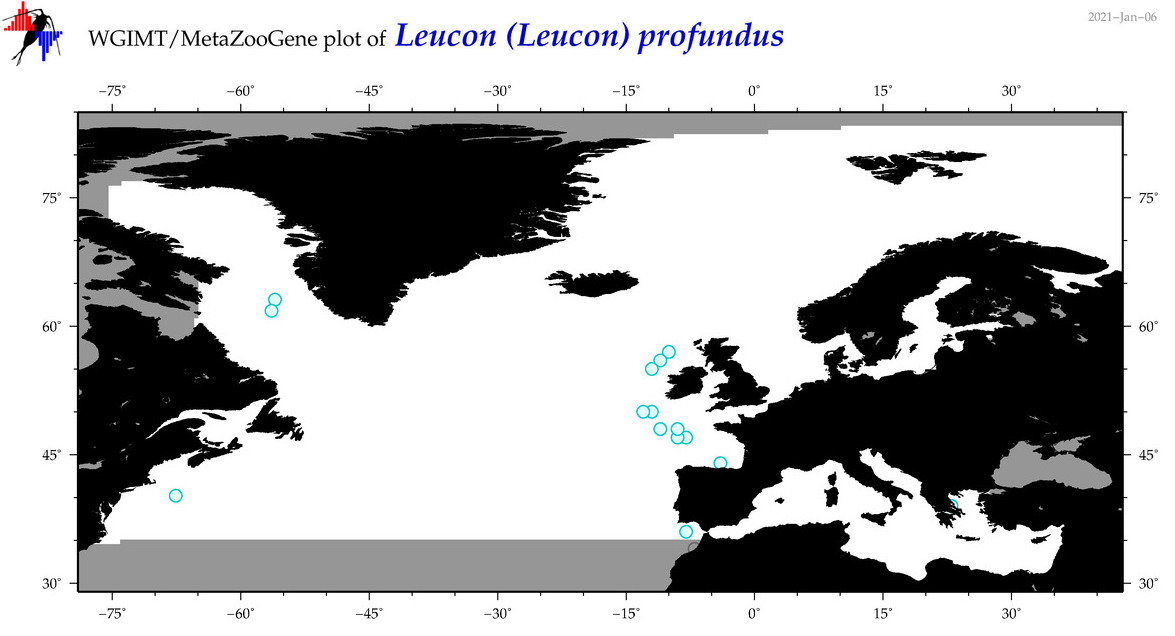 |
accepted
T4094866
R:1:1:0:0 |
| Leucon (Macrauloleucon) siphonatus |
Species
(36) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4091499
R:1:1:0:0 |
| Leucon (Macrauloleucon) spinulosus |
Species
(37) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
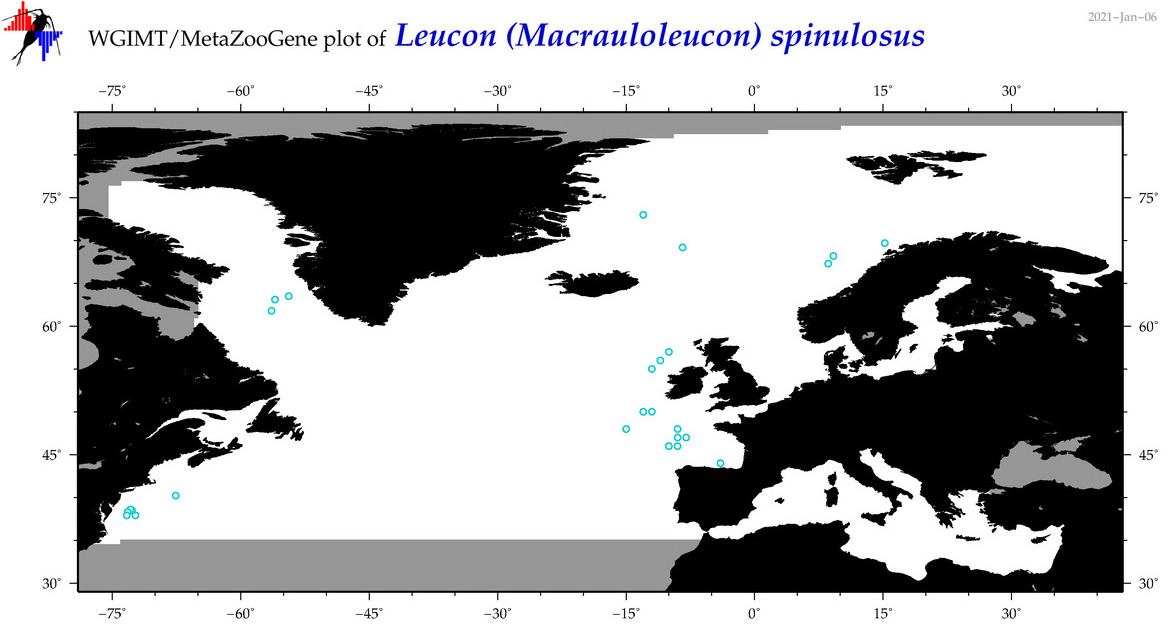 |
accepted
T4091500
R:1:1:0:0 |
| Platycuma holti |
Species
(38) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
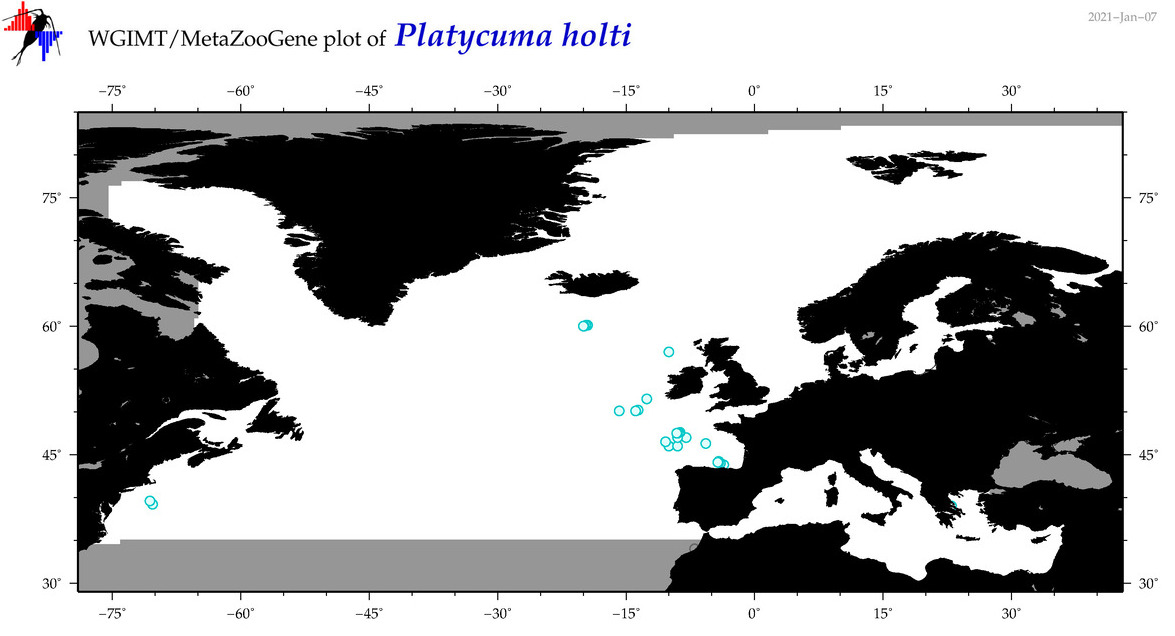 |
accepted
T4064207
R:1:0:0:0 |
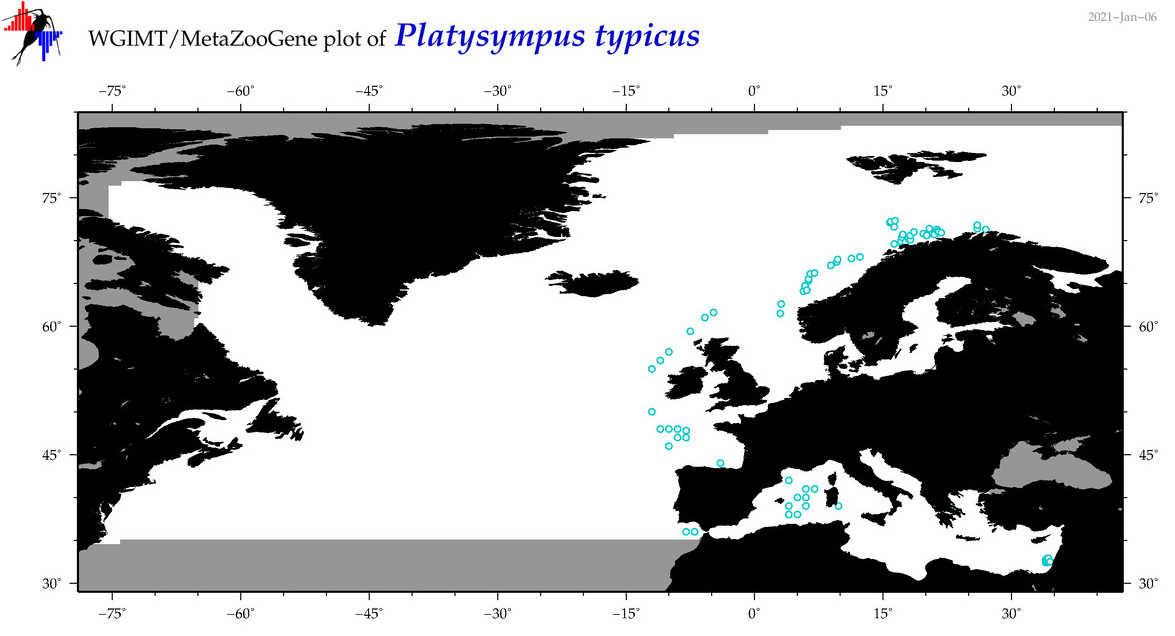
| Platysympus typicus |
Species
(39) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 1
18S = 0
28S = 0
|
 |
accepted
T4064264
R:1:0:0:0 |
| Platytyphlops semiornatus |
Species
(40) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
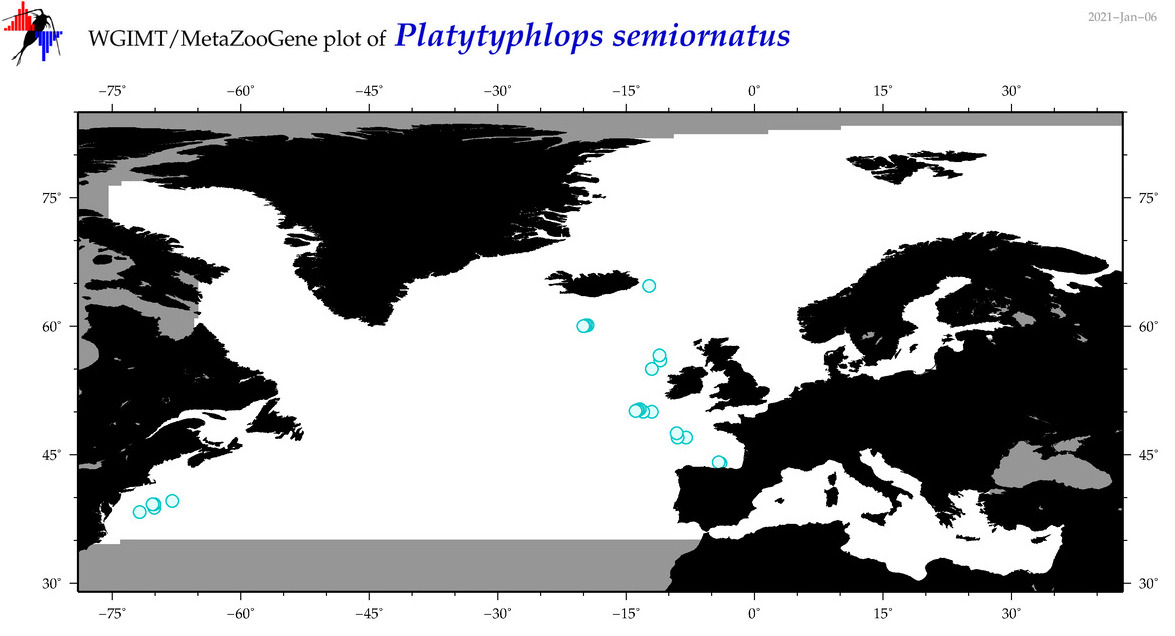 |
accepted
T4064284
R:1:0:0:0 |
| Platytyphlops tuberculatus |
Species
(41) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4064285
R:1:0:0:0 |
| Procampylaspis macronyx |
Species
(42) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
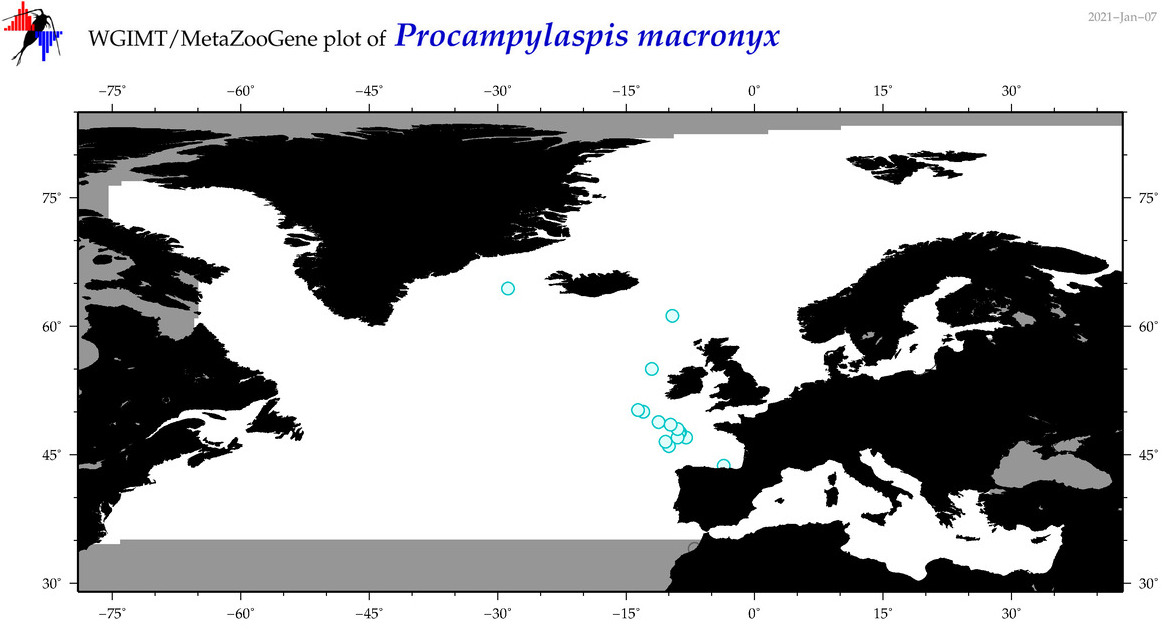 |
accepted
T4065374
R:1:0:0:0 |
| Procampylaspis ommidion |
Species
(43) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
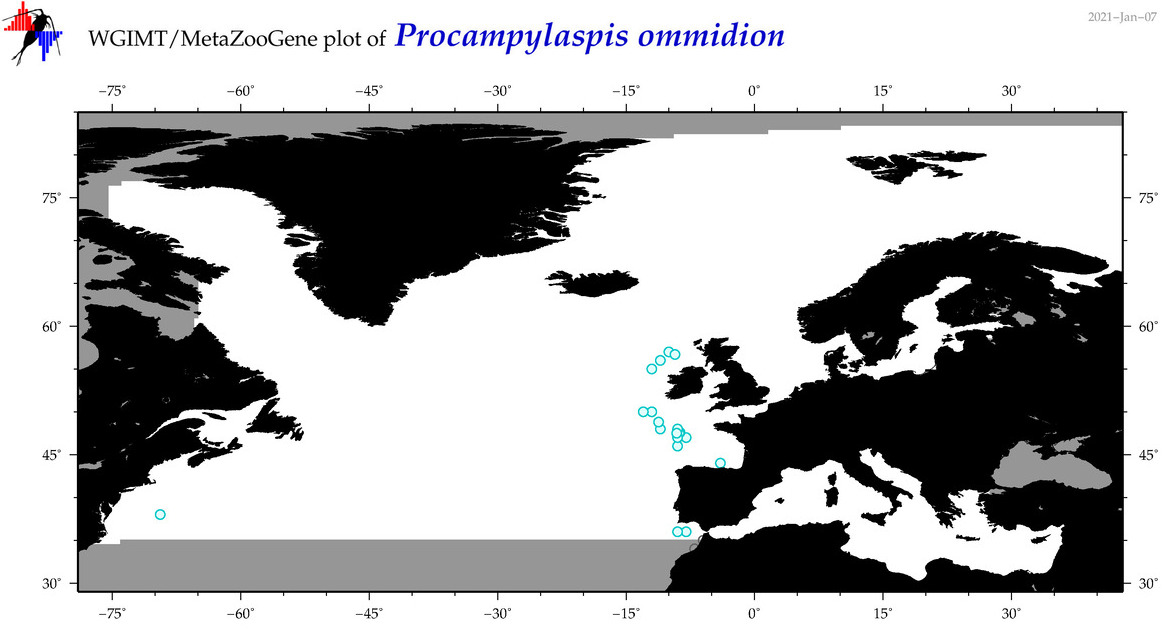 |
accepted
T4065376
R:1:0:0:0 |
| Schizocuma spinoculatum |
Species
(44) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
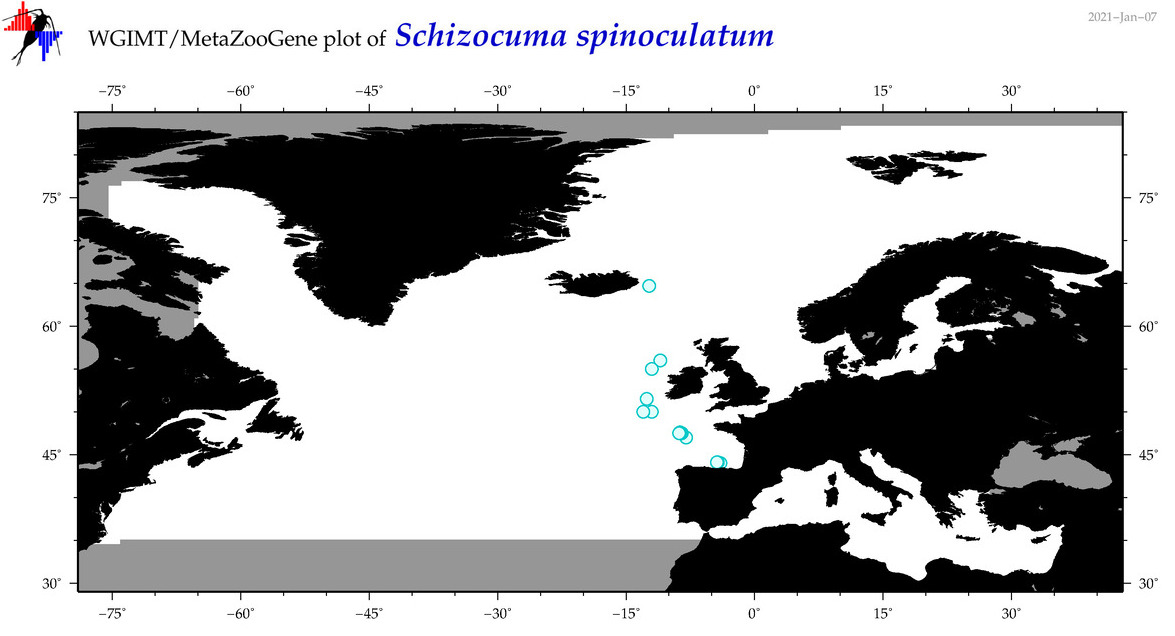 |
accepted
T4067382
R:1:0:0:0 |
| Styloptocuma erectum |
Species
(45) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
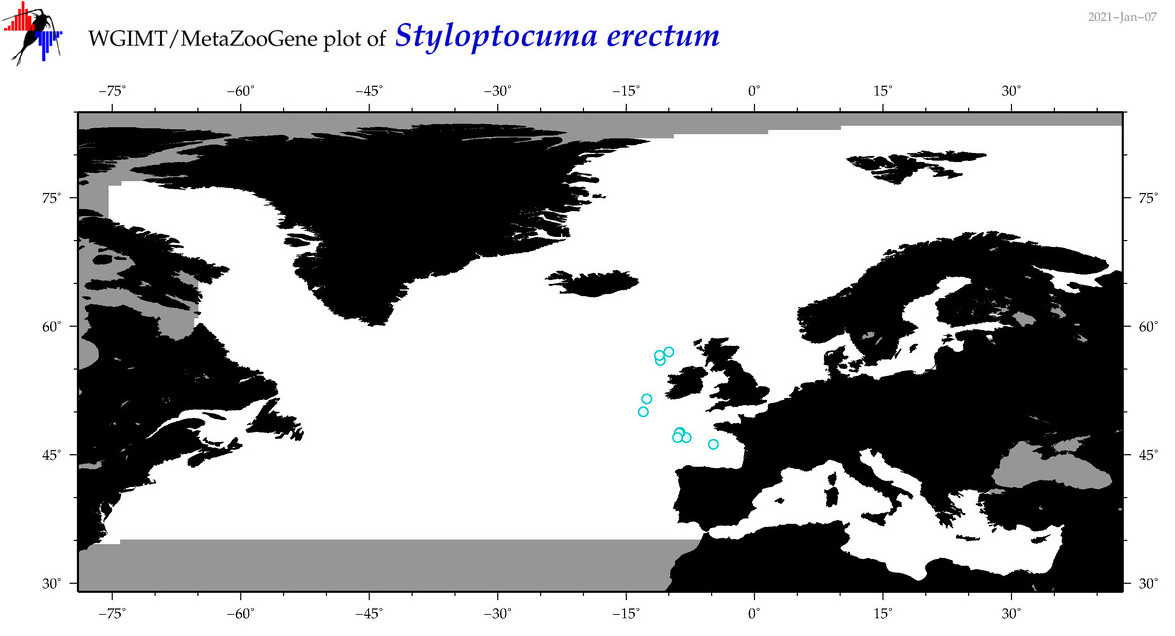 |
accepted
T4069392
R:1:0:0:0 |
| Styloptocuma gracillimum |
Species
(46) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
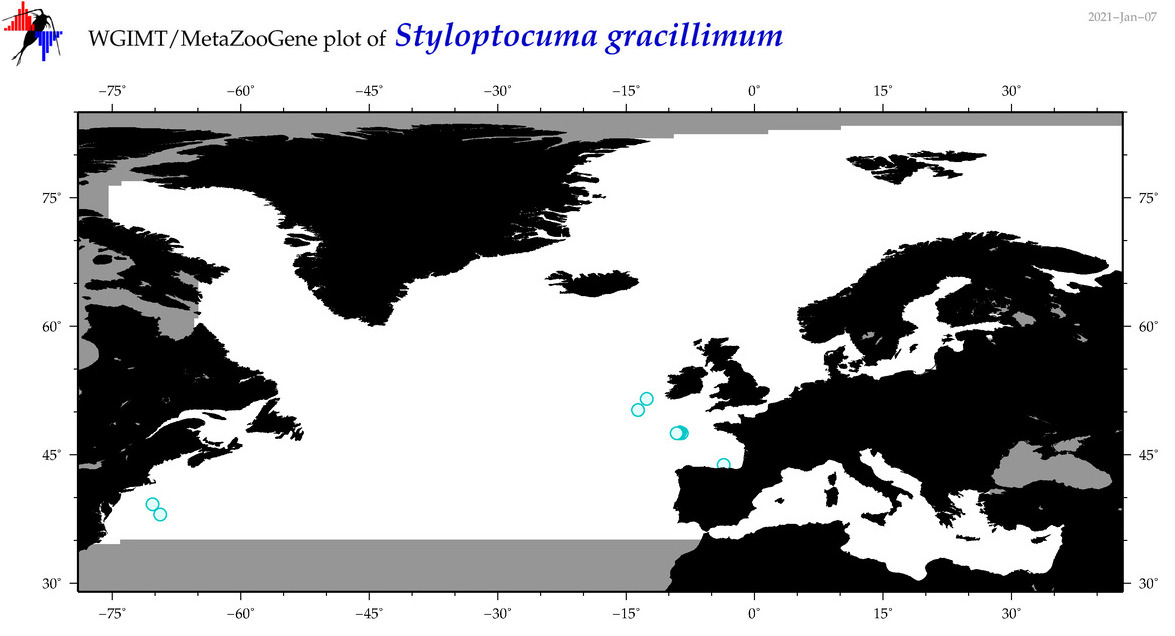 |
accepted
T4069396
R:1:0:0:0 |