Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-26 |
| Acanthomysis longicornis |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
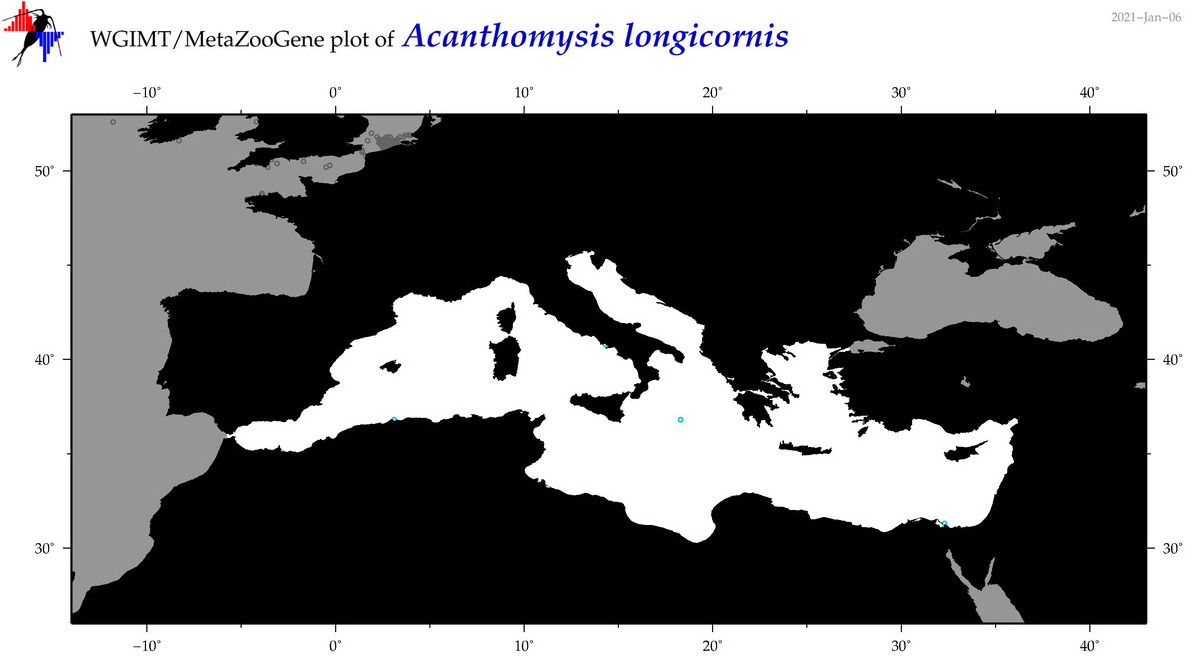 |
accepted
T4007118
R:1:0:0:0 |
| Anchialina agilis |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
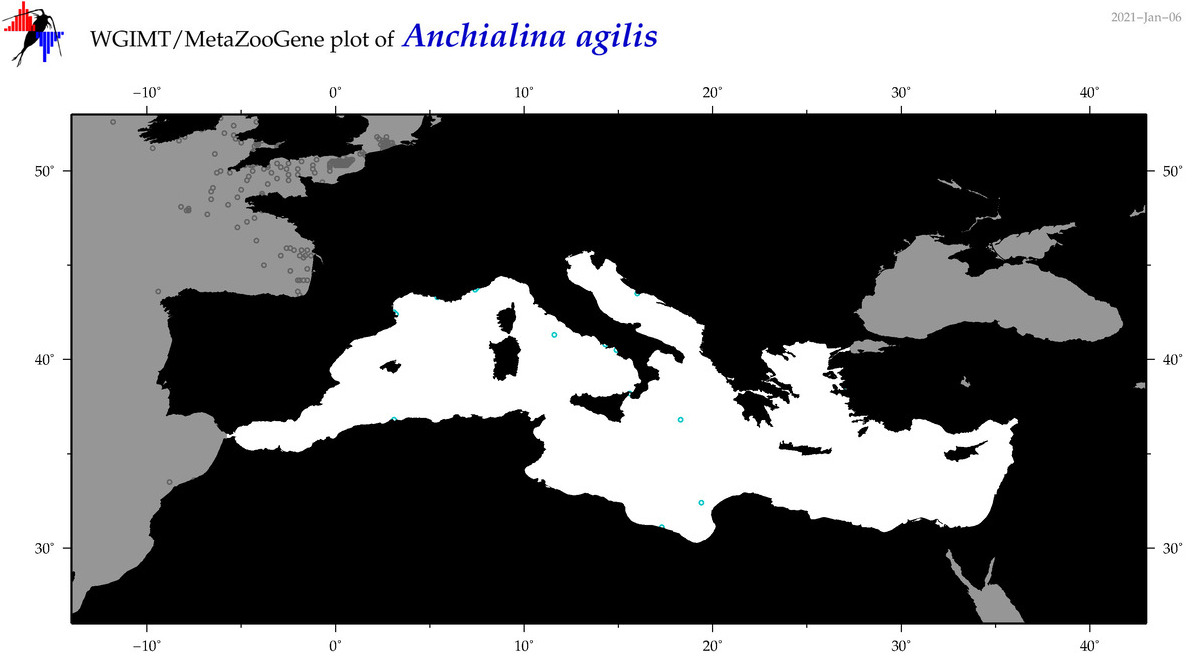 |
accepted
T4007122
R:1:0:0:0 |
| Anchialina oculata |
Species
(3) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4043363
R:1:0:0:0 |
| Arachnomysis leuckartii |
Species
(4) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
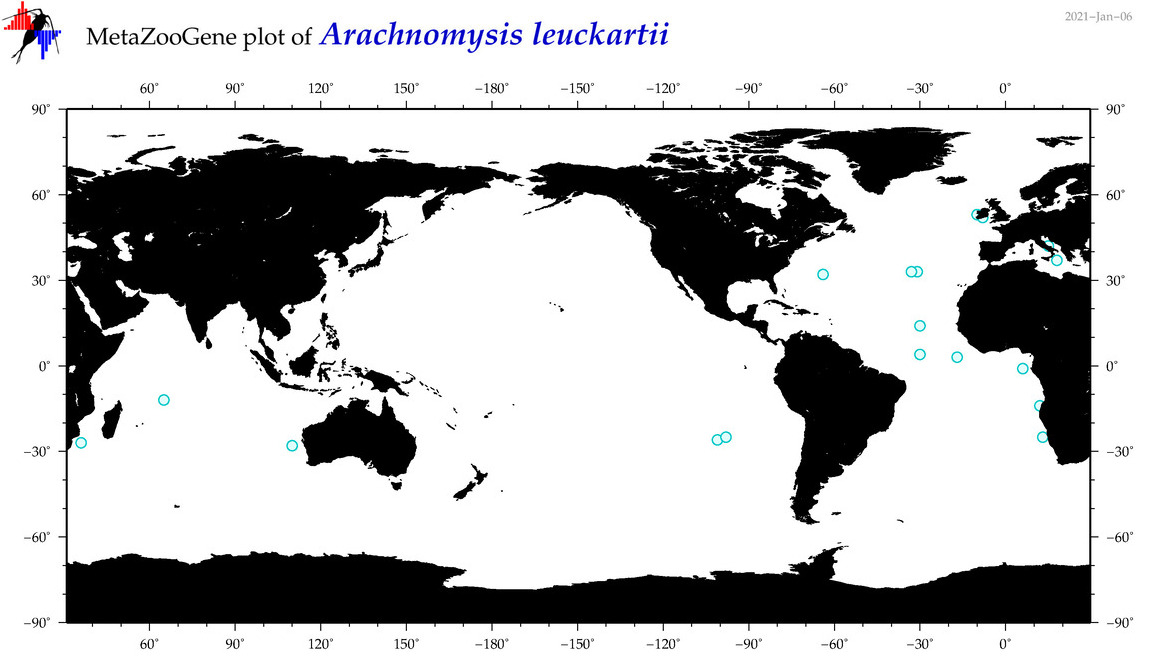 |
accepted
T4044222
R:1:0:0:0 |
| Boreomysis (Boreomysis) arctica |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
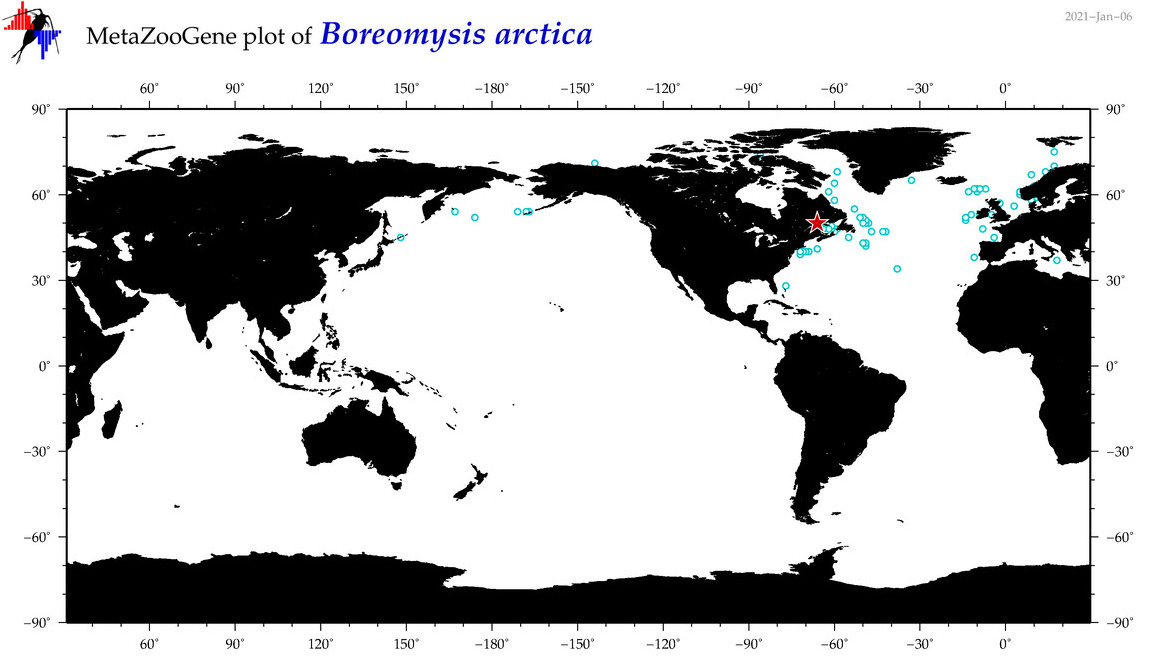 |
accepted
T4007130
R:1:0:0:0 |
| Boreomysis (Petryashovia) megalops |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4007131
R:1:0:0:0 |
| Calyptomma puritani |
Species
(7) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4046429
R:1:0:0:0 |
| Dactylamblyops corberai |
Species
(8) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4049211
R:1:0:0:0 |
| Diamysis bahirensis |
Species
(9) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
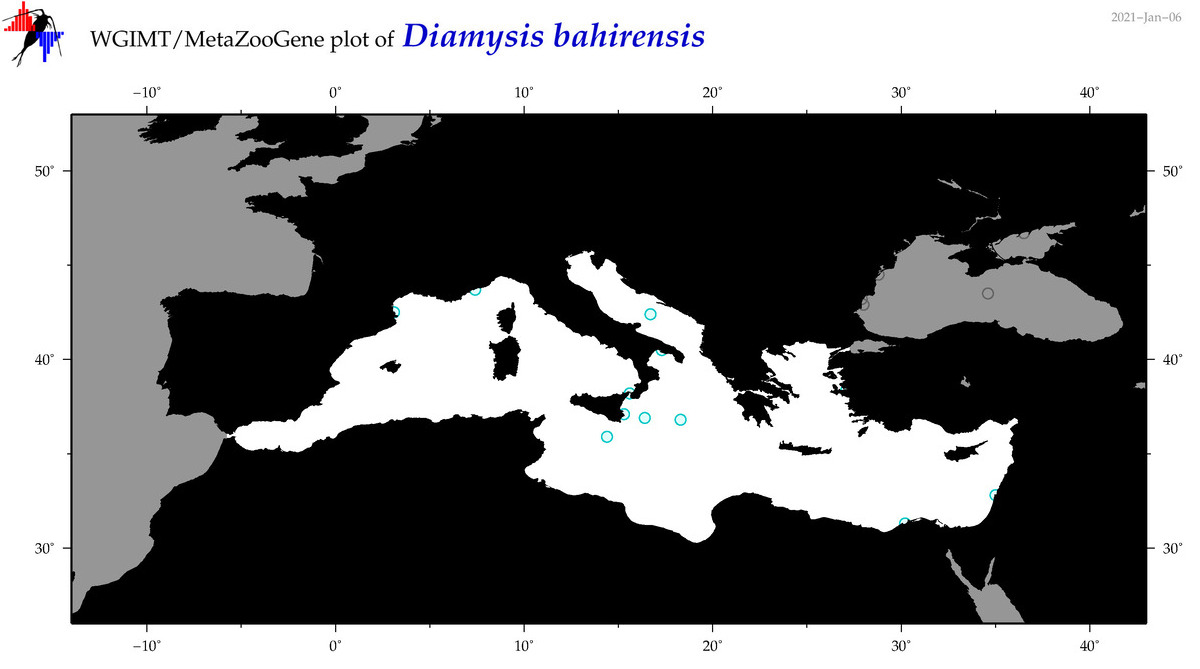 |
accepted
T4049559
R:1:1:1:0 |
| Erythrops elegans |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
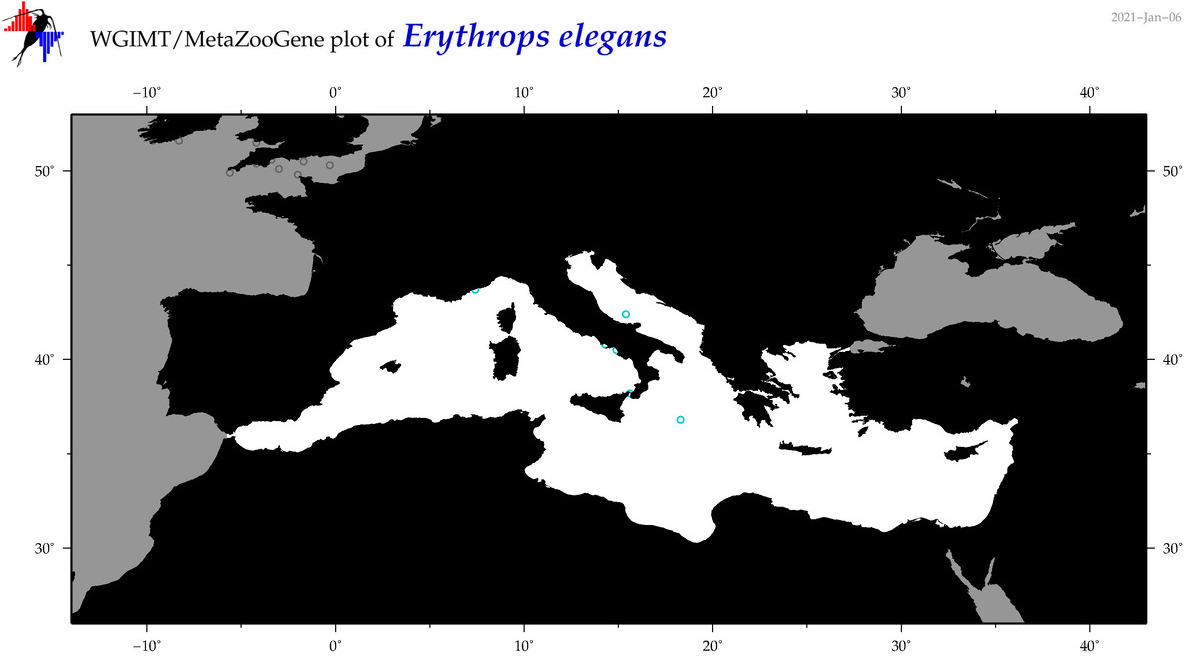 |
accepted
T4007140
R:1:0:0:0 |
| Erythrops erythrophthalmus |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
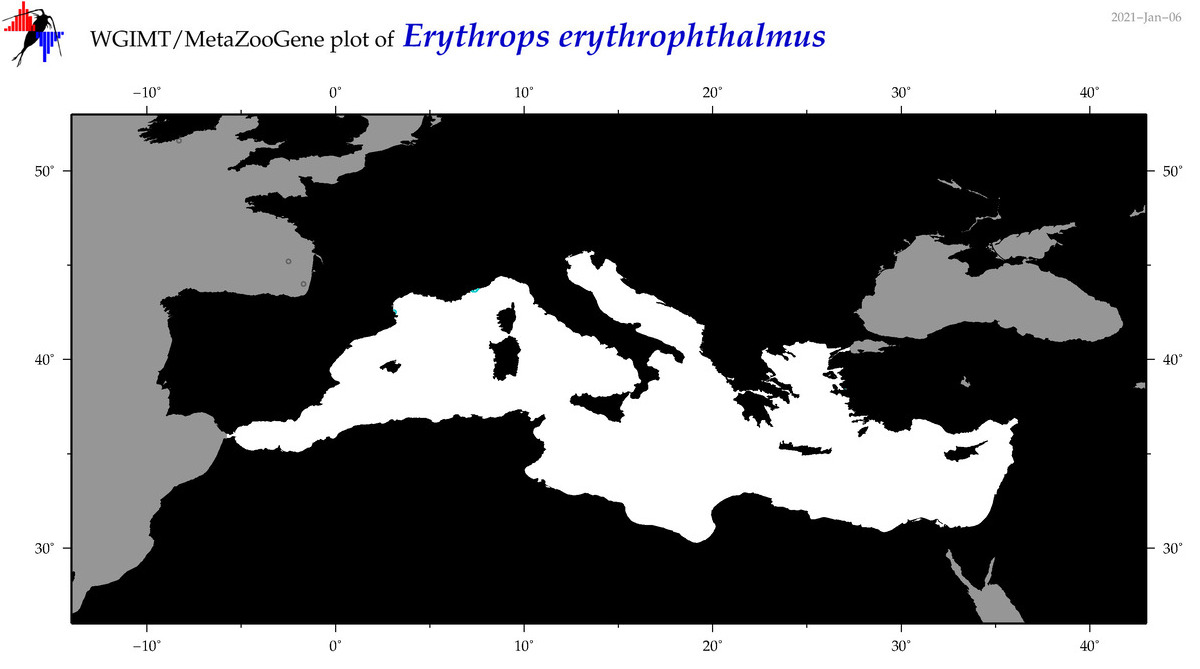 |
accepted
T4001063
R:1:0:0:0 |
| Erythrops neapolitanus |
Species
(12) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
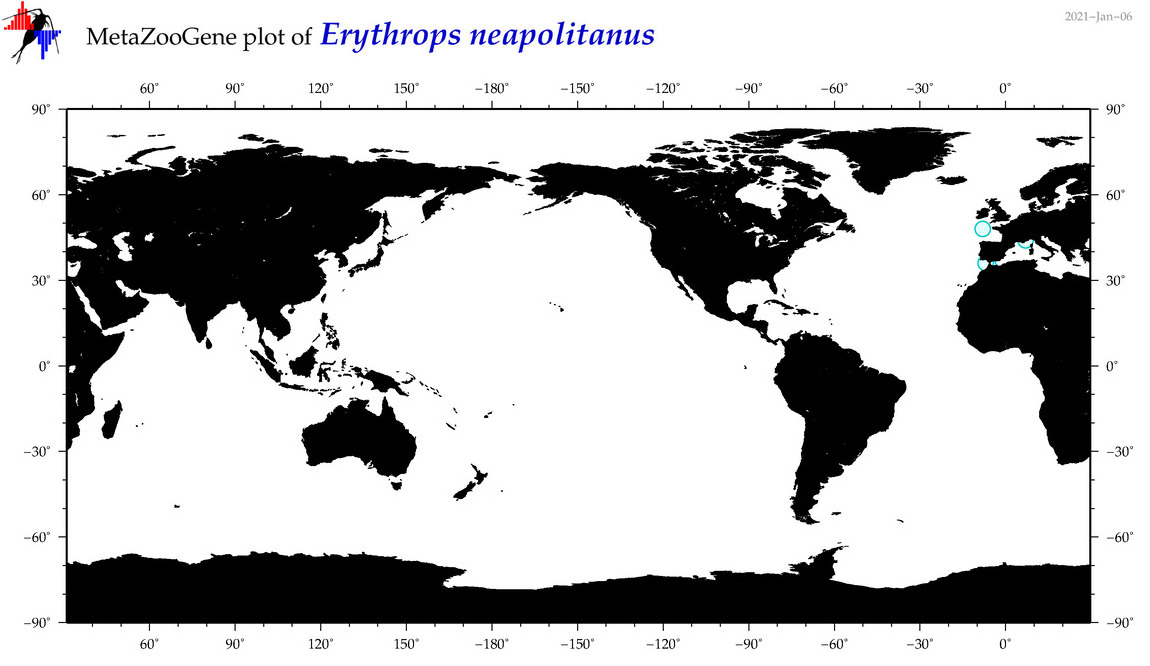 |
accepted
T4051082
R:1:0:0:0 |
| Euchaetomera glyphidophthalmica |
Species
(13) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
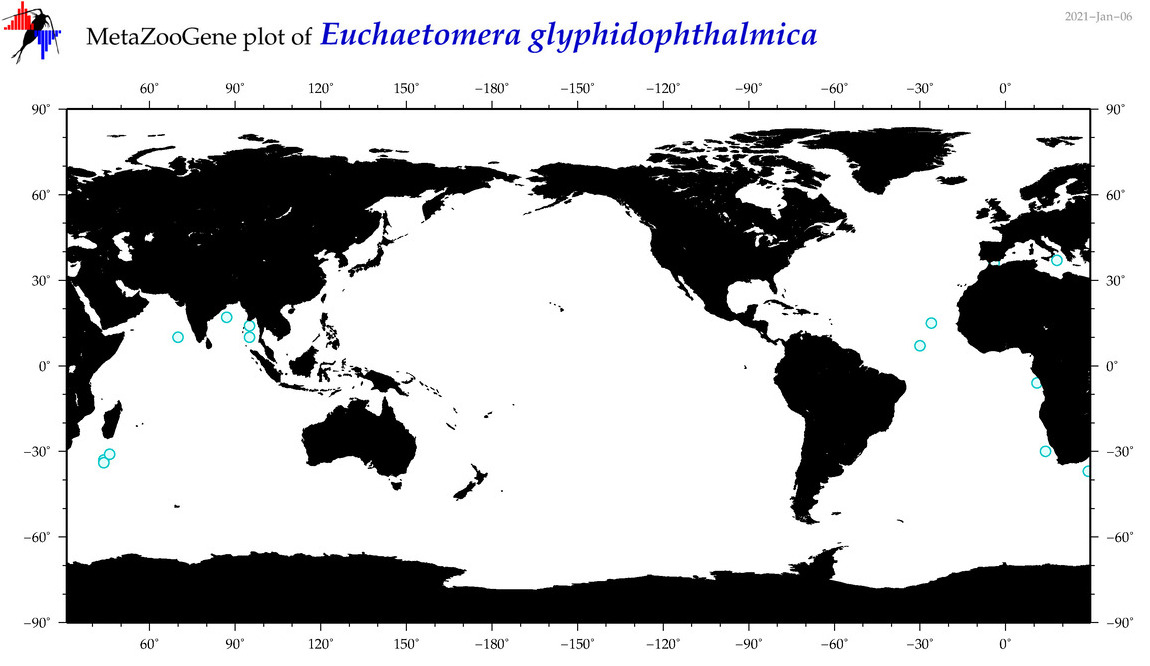 |
accepted
T4051239
R:1:0:0:0 |
| Euchaetomera tenuis |
Species
(14) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
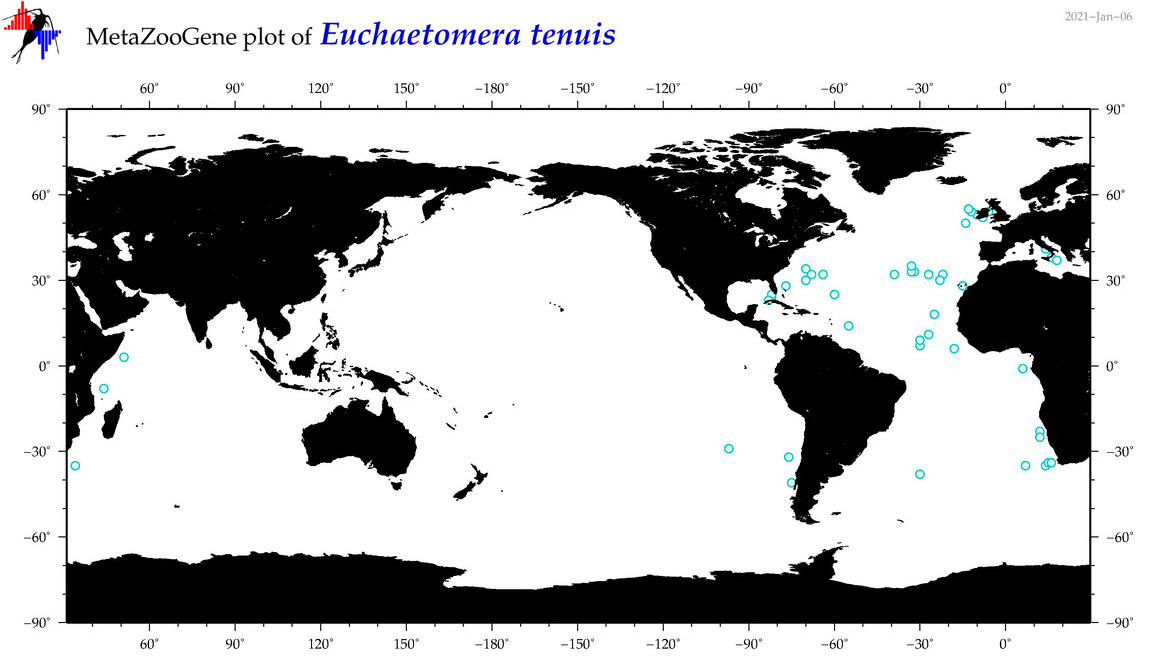 |
accepted
T4007144
R:1:0:0:0 |
| Euchaetomeropsis merolepis |
Species
(15) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4051243
R:1:0:0:0 |
| Gastrosaccus mediterraneus |
Species
(16) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4052597
R:1:0:0:0 |
| Gastrosaccus sanctus |
Species
(17) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
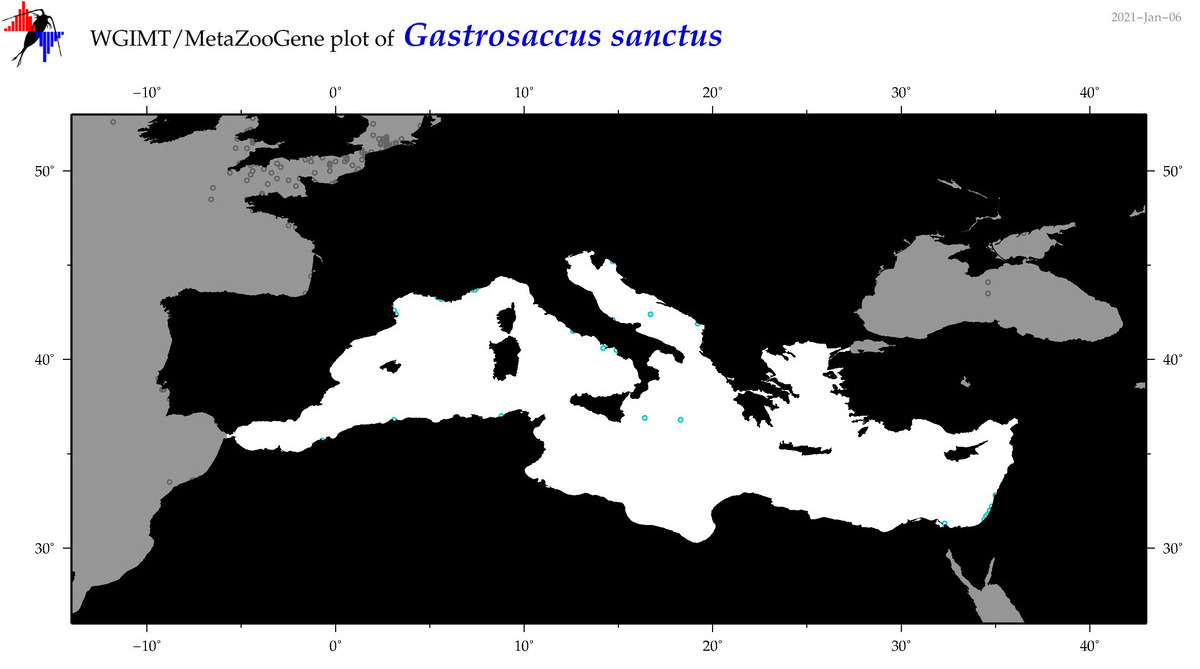 |
accepted
T4007148
R:1:0:0:0 |
| Gastrosaccus spinifer |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
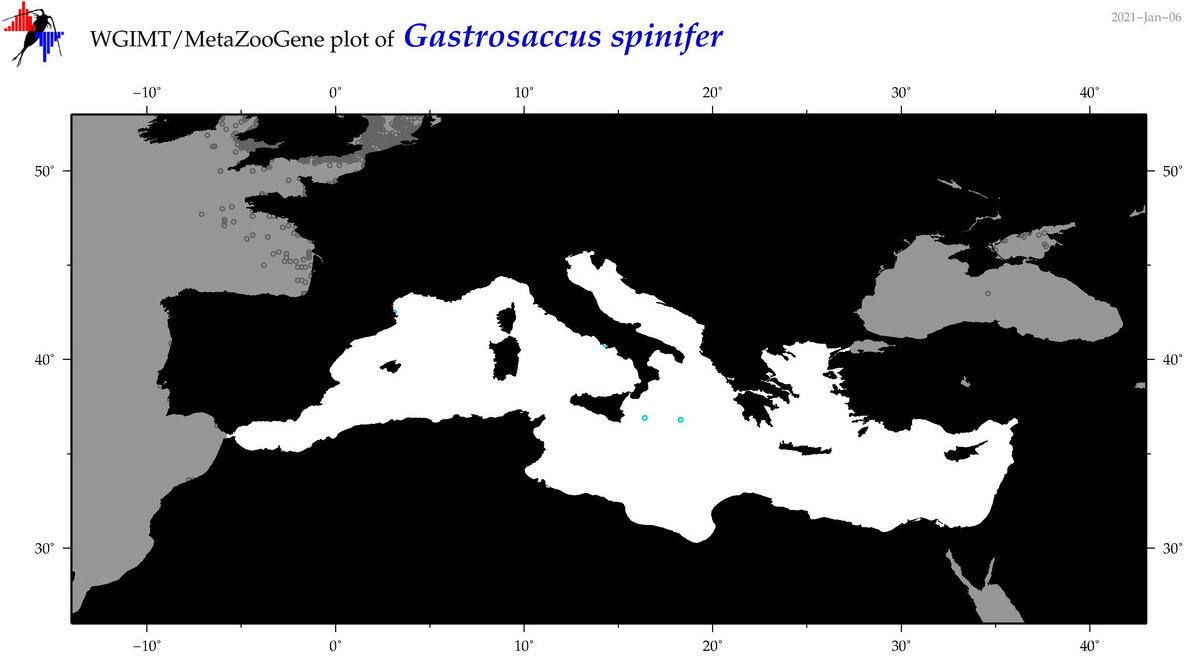 |
accepted
T4007149
R:1:1:0:0 |
| Haplostylus bacescui |
Species
(19) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4053563
R:1:0:0:0 |
| Haplostylus lobatus |
Species
(20) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
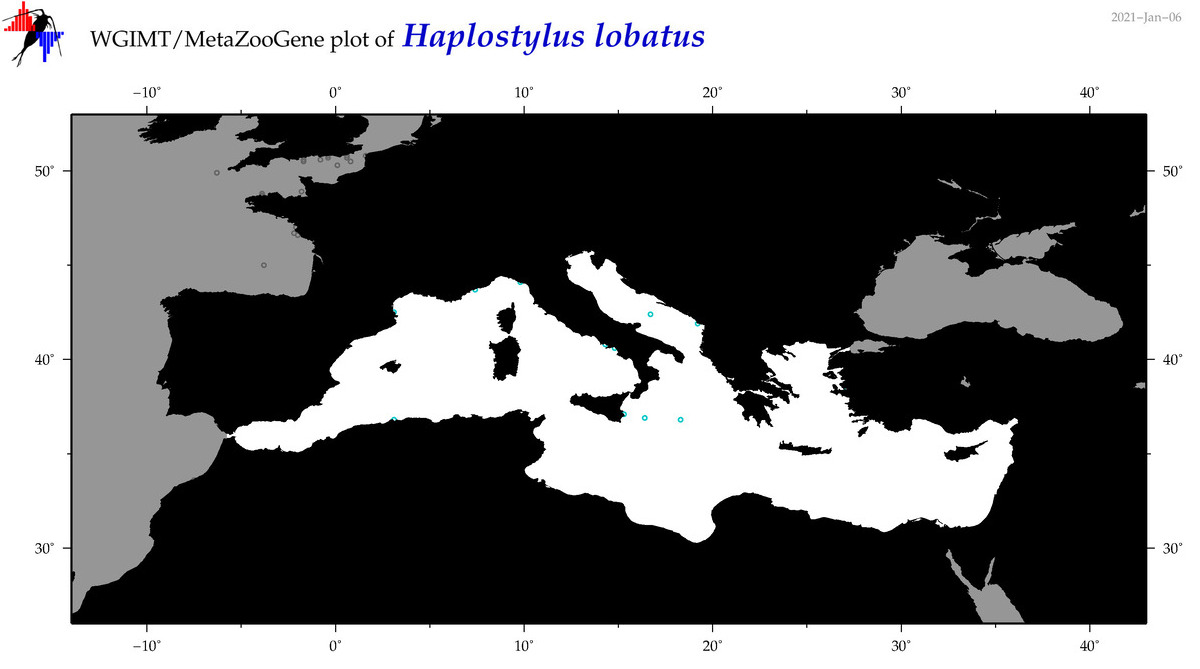 |
accepted
T4007152
R:1:0:0:0 |
| Haplostylus magnilobatus |
Species
(21) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4053568
R:1:0:0:0 |
| Haplostylus normani |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
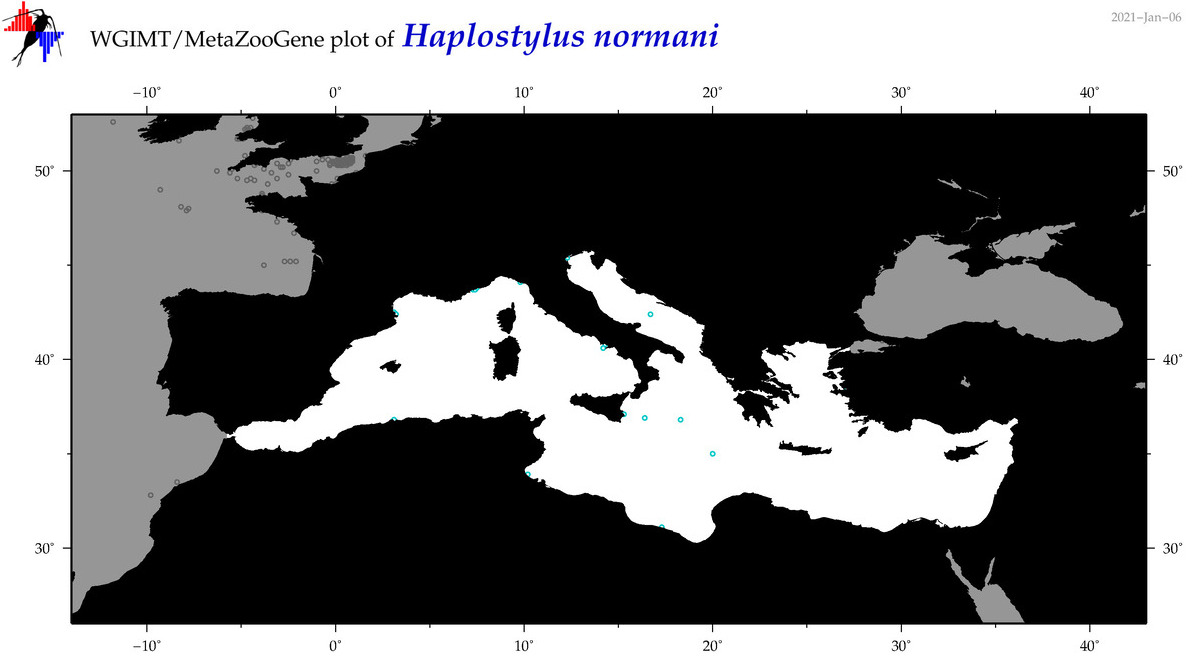 |
accepted
T4007153
R:1:0:0:0 |
| Hemimysis abyssicola |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
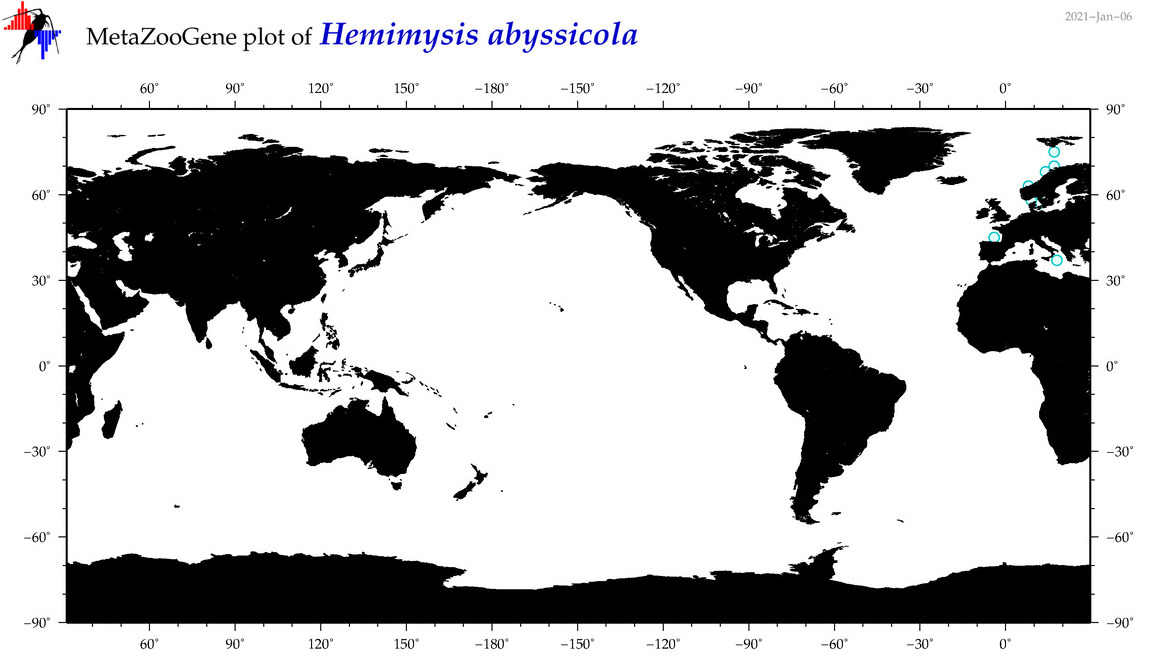 |
accepted
T4053856
R:1:0:0:0 |
| Hemimysis lamornae |
Species
(24) |
COI
12S
16S
18S
28S
|
COI = 33
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
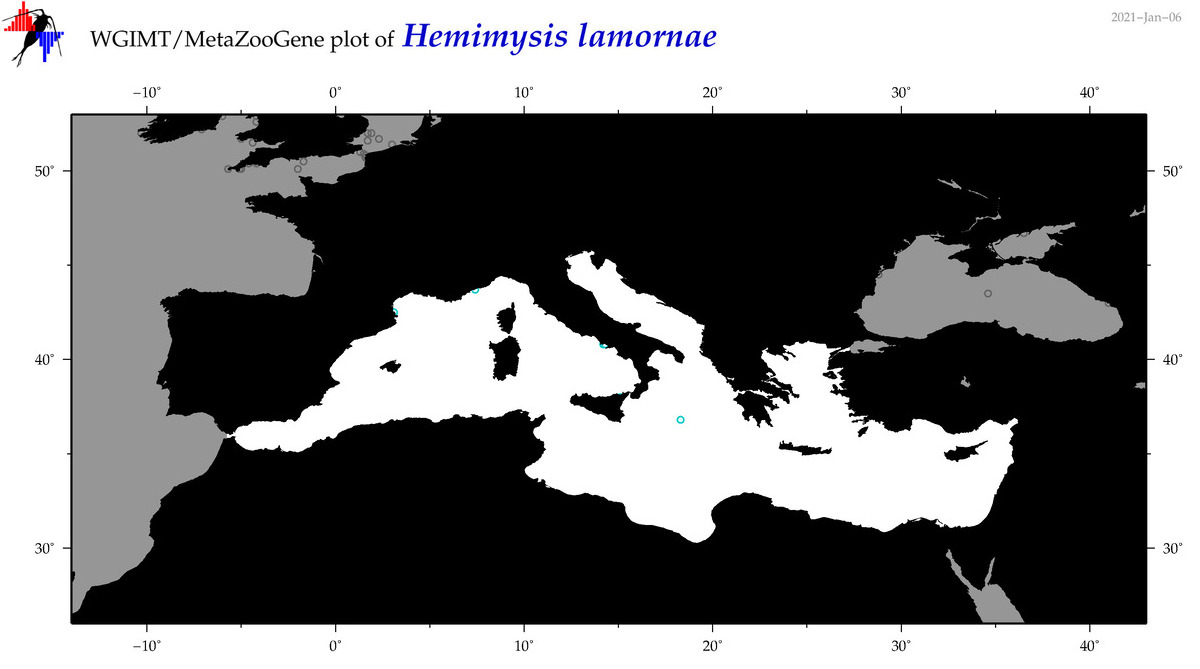 |
accepted
T4007155
R:1:0:0:0 |
| Hemimysis speluncola |
Species
(25) |
COI
12S
16S
18S
28S
|
COI = 15
12S = 0
16S = 1
18S = 2
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
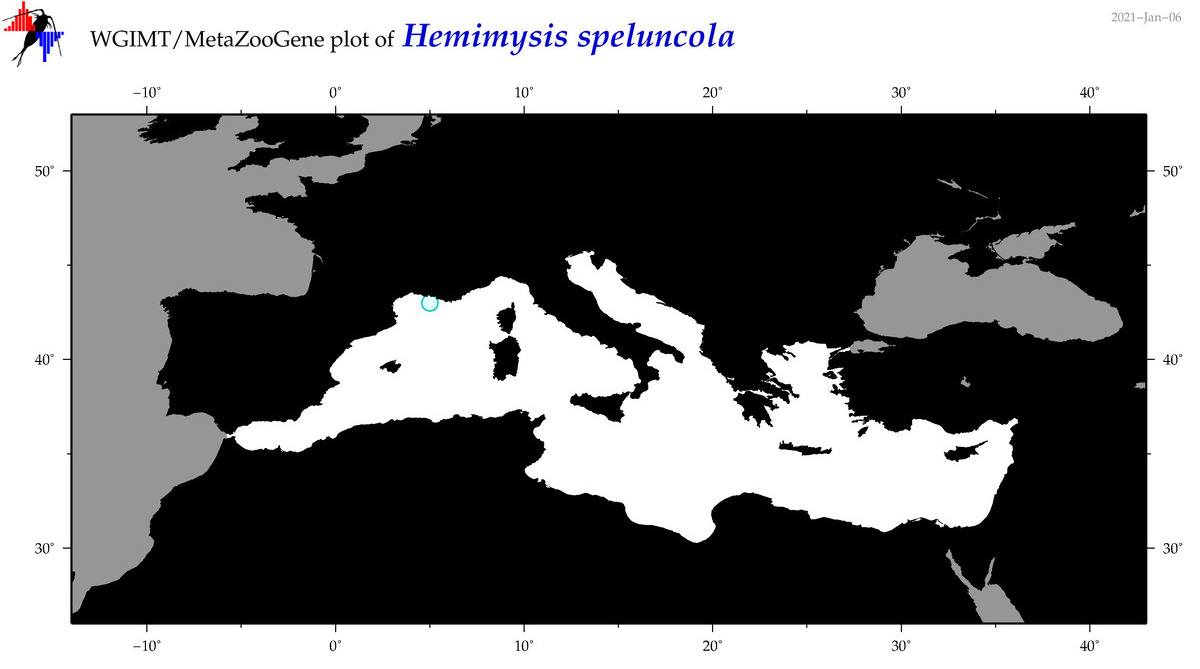 |
accepted
T4012093
R:1:0:0:0 |
| Heteromysis (Heteromysis) arianii |
Species
(26) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4094521
R:1:0:0:0 |
| Heteromysis (Heteromysis) formosa |
Species
(27) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
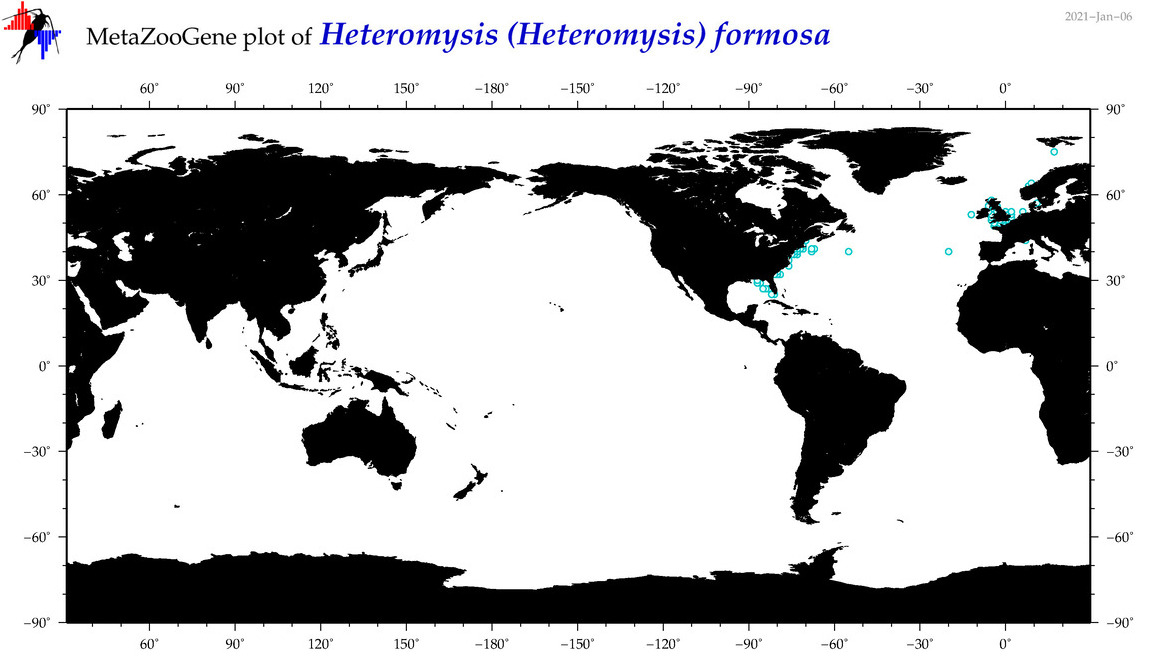 |
accepted
T4007215
R:1:0:0:0 |
| Heteromysis (Heteromysis) microps |
Species
(28) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
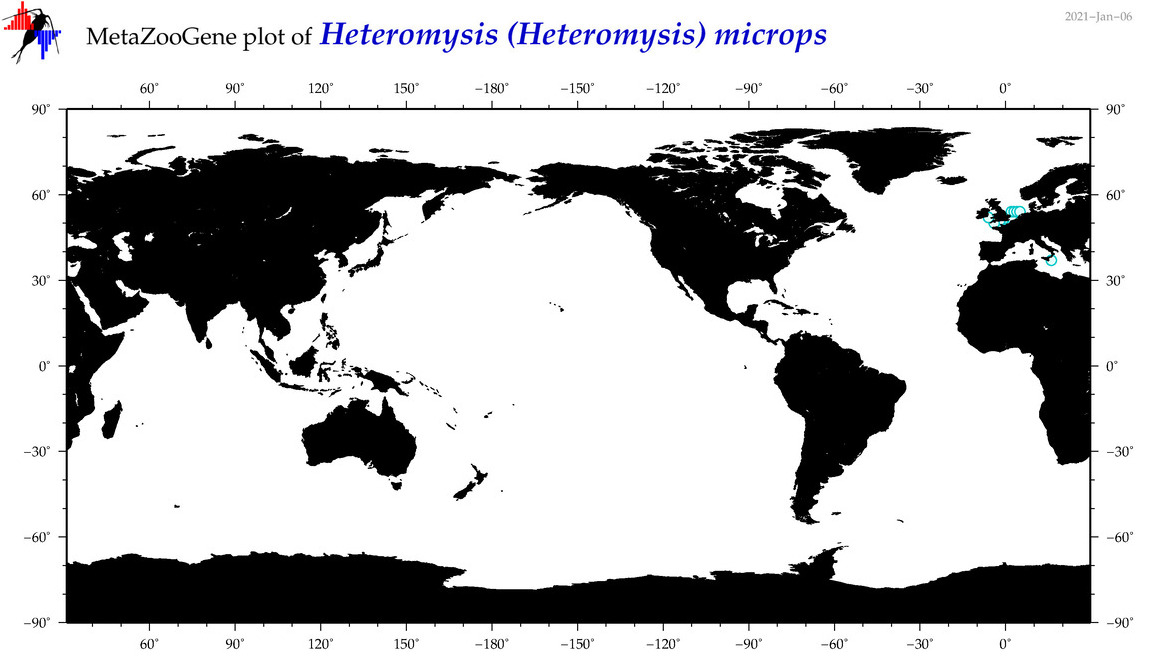 |
accepted
T4007216
R:1:0:0:0 |
| Hypererythrops zimmeri |
Species
(29) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
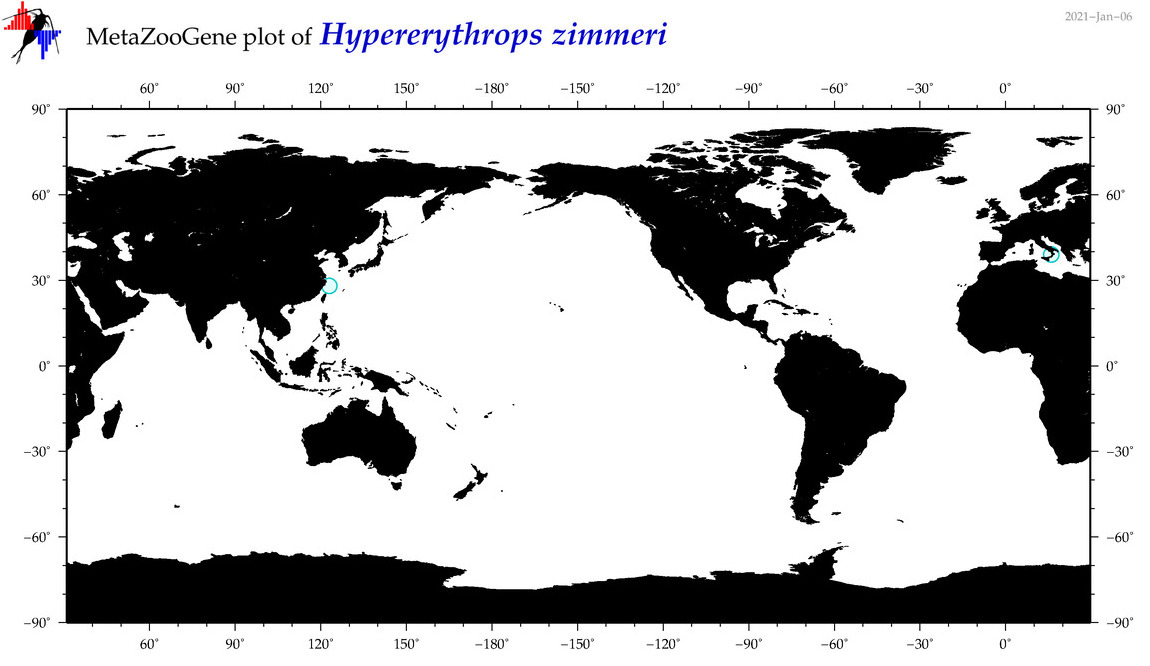 |
accepted
T4054603
R:1:1:0:0 |
| Leptomysis buergii |
Species
(30) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4056191
R:1:0:0:0 |
| Leptomysis gracilis |
Species
(31) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4007163
R:1:0:0:0 |
| Leptomysis lingvura |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 2
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
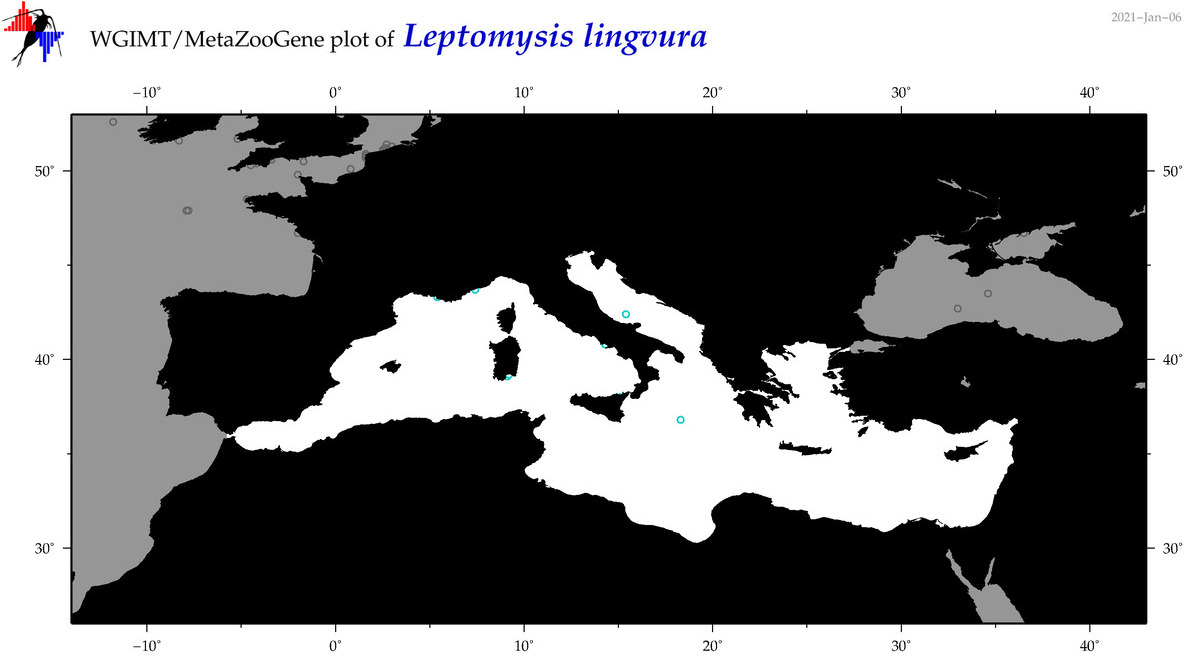 |
accepted
T4007164
R:1:0:0:0 |
| Leptomysis mediterranea |
Species
(33) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
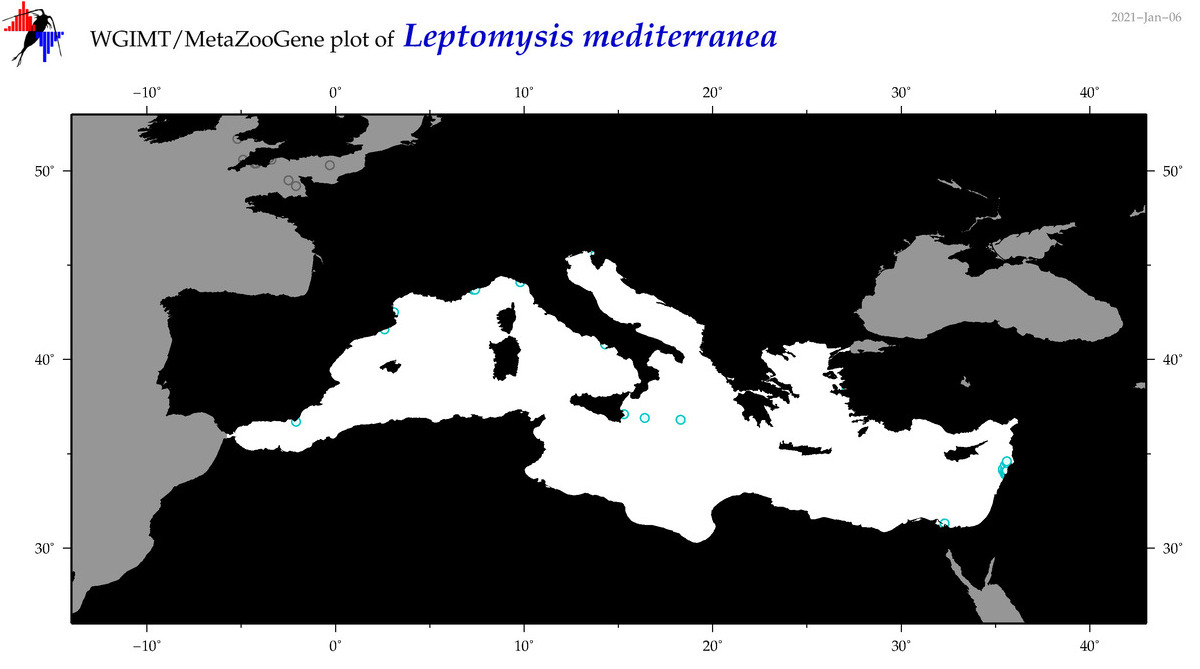 |
accepted
T4007165
R:1:1:0:0 |
| Leptomysis megalops |
Species
(34) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
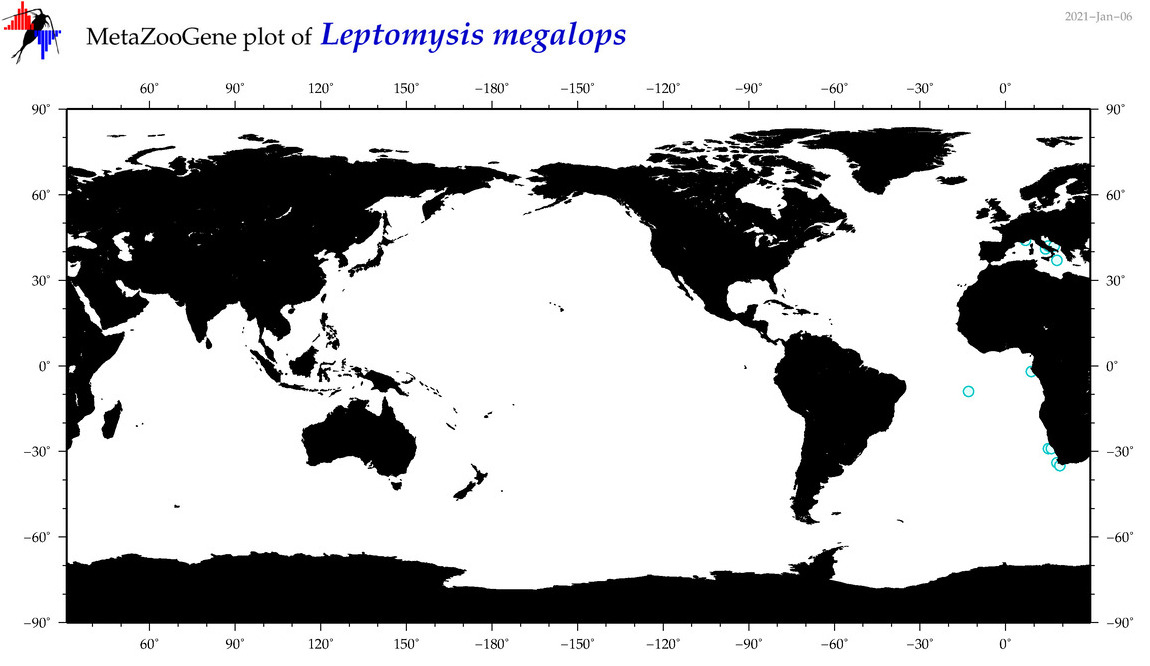 |
accepted
T4056194
R:1:0:0:0 |
| Leptomysis posidoniae |
Species
(35) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4056195
R:1:0:0:0 |
| Leptomysis truncata |
Species
(36) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
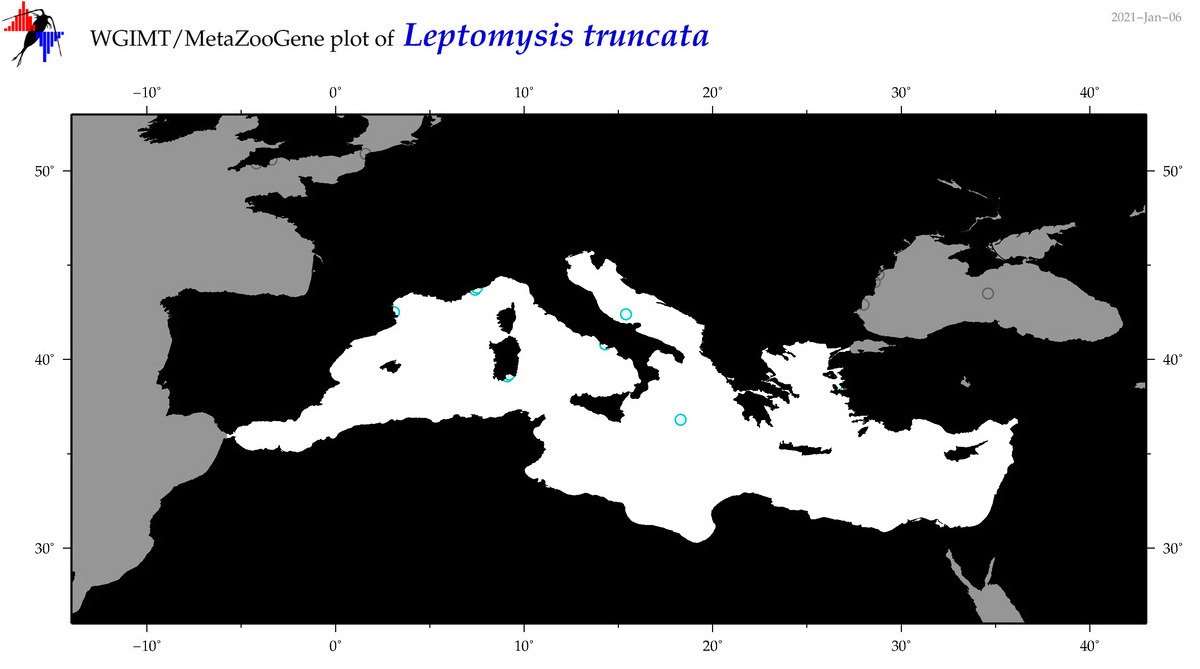 |
accepted
T4007166
R:1:0:0:0 |
| Mesopodopsis aegyptia |
Species
(37) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
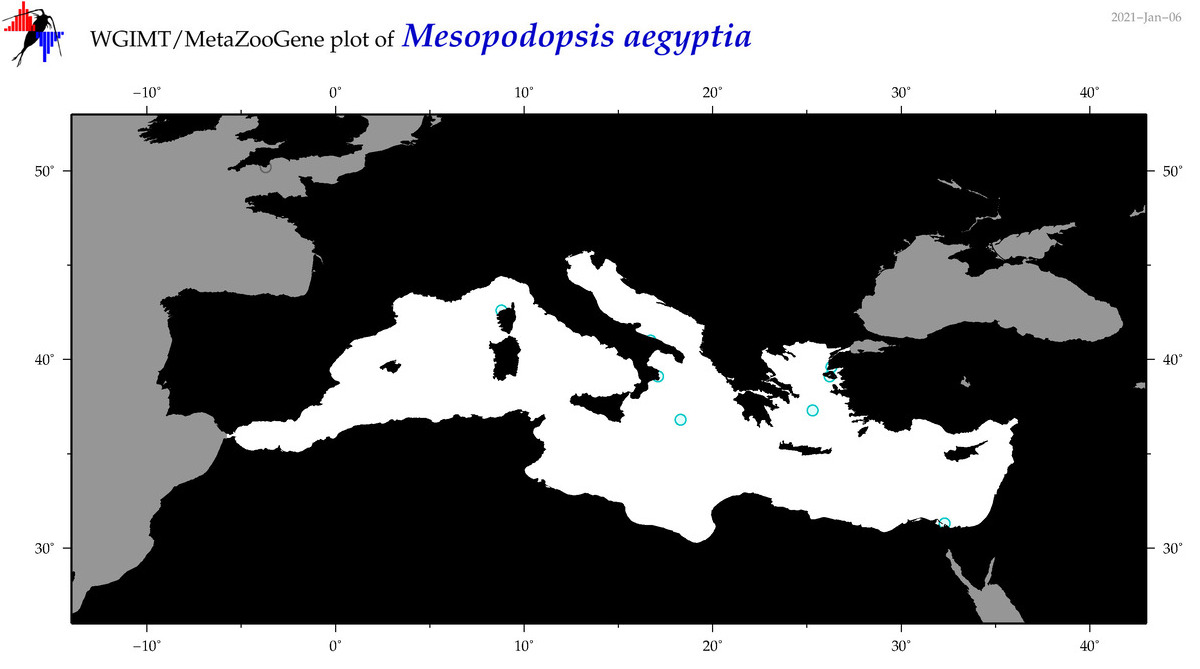 |
accepted
T4058116
R:1:1:0:0 |
| Mesopodopsis slabberi |
Species
(38) |
COI
12S
16S
18S
28S
|
COI = 86
12S = 0
16S = 17
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
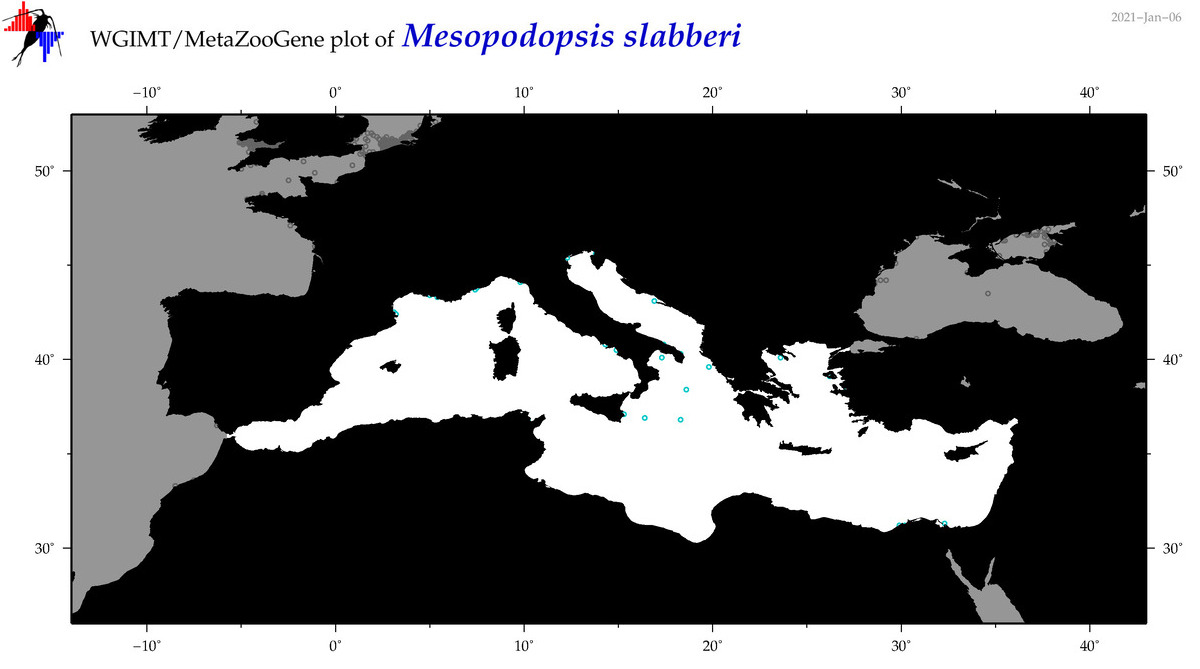 |
accepted
T4007168
R:1:1:0:0 |
| Mysideis parva |
Species
(39) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
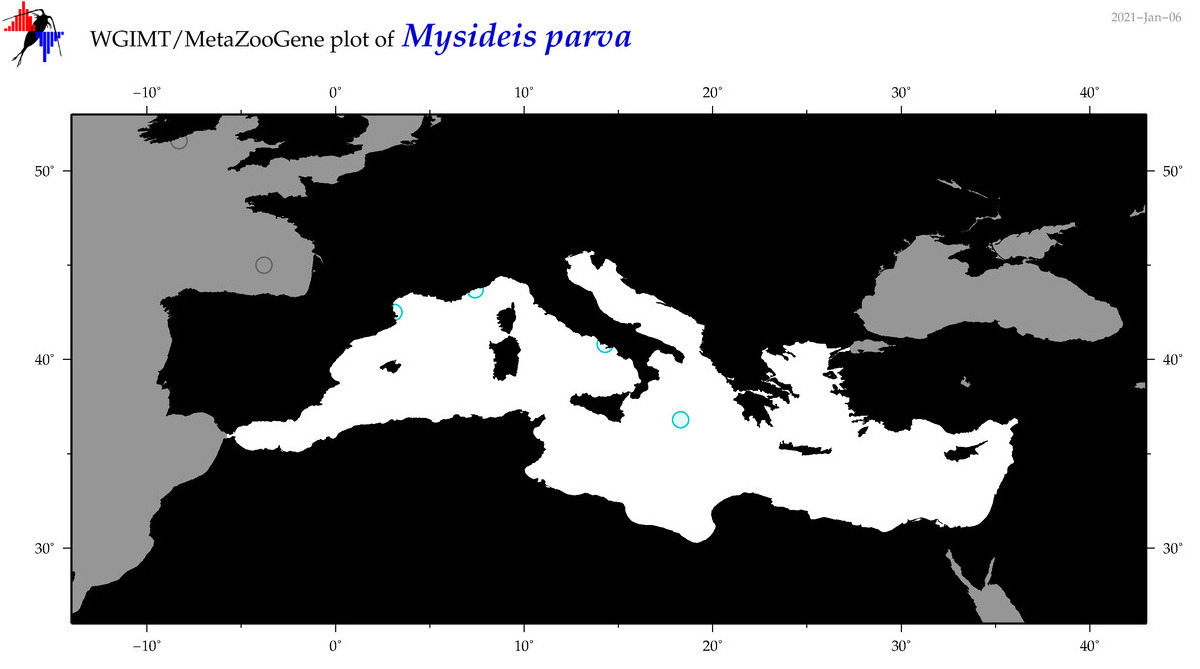 |
accepted
T4059602
R:1:0:0:0 |
| Mysidella typica |
Species
(40) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4059608
R:1:0:0:0 |
| Mysidetes farrani |
Species
(41) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4059614
R:1:0:0:0 |
| Mysidopsis angusta |
Species
(42) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
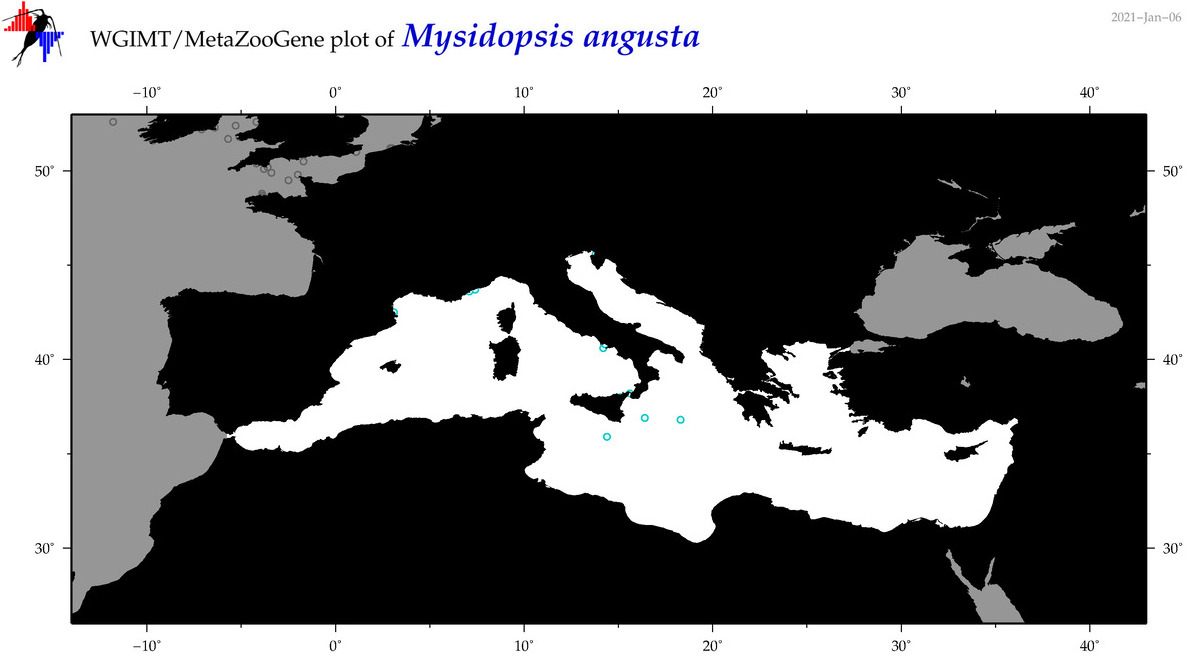 |
accepted
T4007173
R:1:0:0:0 |
| Mysidopsis didelphys |
Species
(43) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
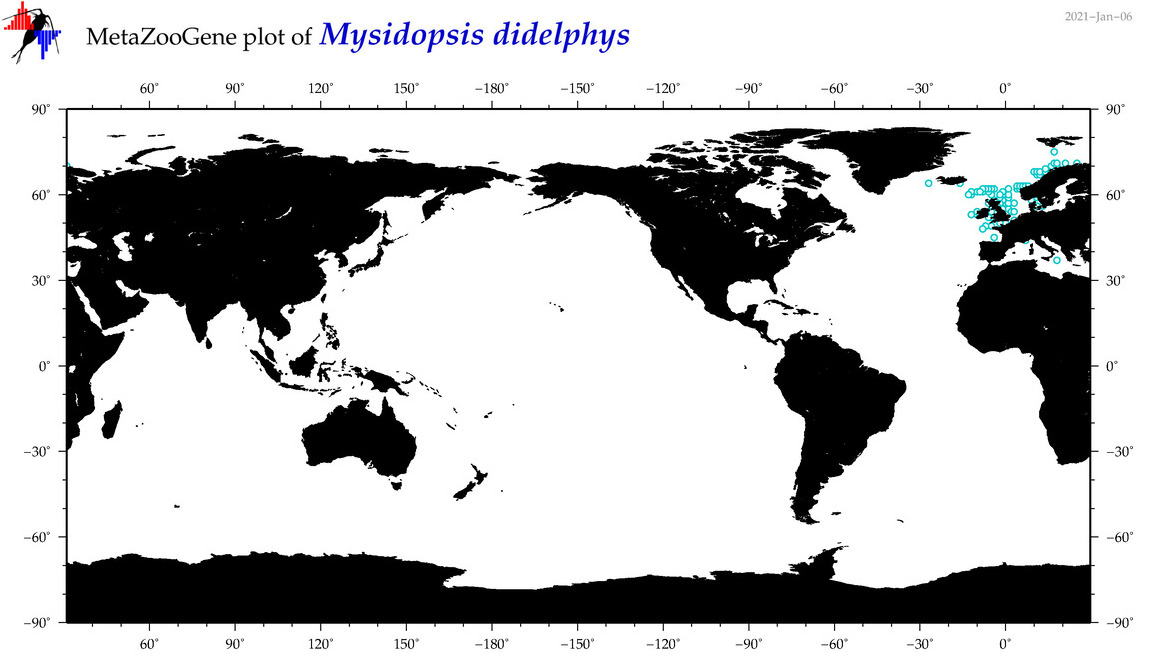 |
accepted
T4007174
R:1:0:0:0 |
| Mysidopsis gibbosa |
Species
(44) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
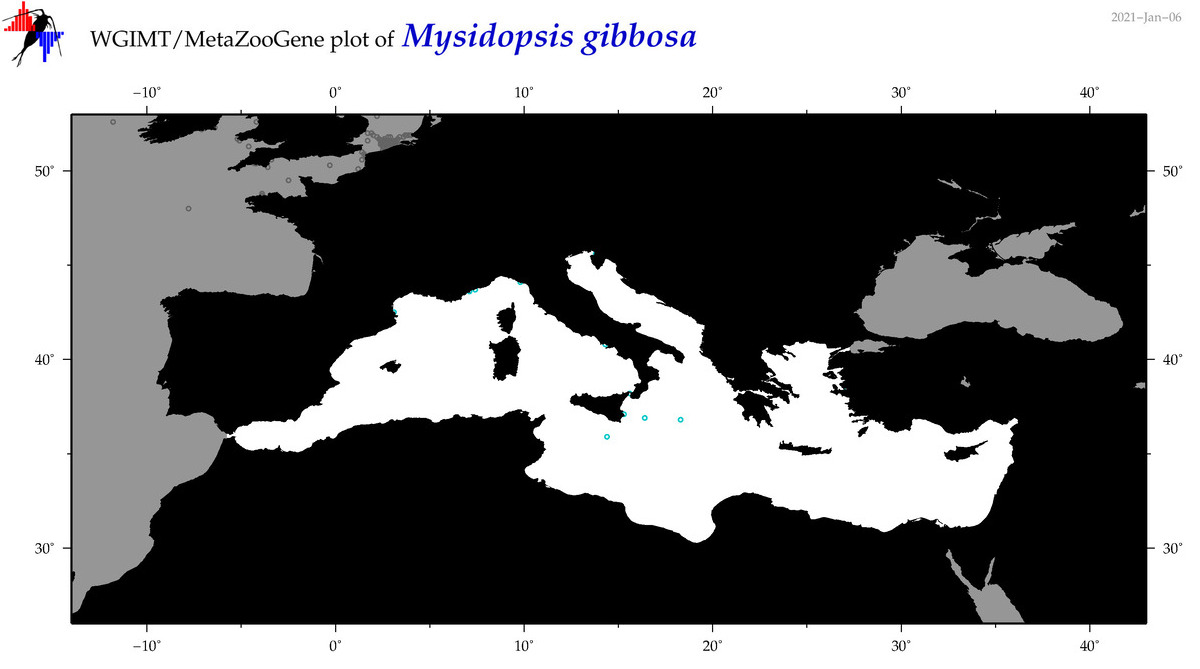 |
accepted
T4007176
R:1:0:0:0 |
| Mysis amblyops |
Species
(45) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 2
18S = 1
28S = 0
|
COI = 1
12S = 0
16S = 2
18S = 1
28S = 0
|
 |
accepted
T4013197
R:1:0:0:0 |
| Mysis caspia |
Species
(46) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 2
18S = 1
28S = 0
|
COI = 1
12S = 0
16S = 2
18S = 1
28S = 0
|
 |
accepted
T4013198
R:1:0:0:0 |
| Mysis microphthalma |
Species
(47) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 1
18S = 1
28S = 0
|
COI = 3
12S = 0
16S = 1
18S = 1
28S = 0
|
 |
accepted
T4013201
R:1:0:0:0 |
| Mysis relicta |
Species
(48) |
COI
12S
16S
18S
28S
|
COI = 31
12S = 0
16S = 2
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
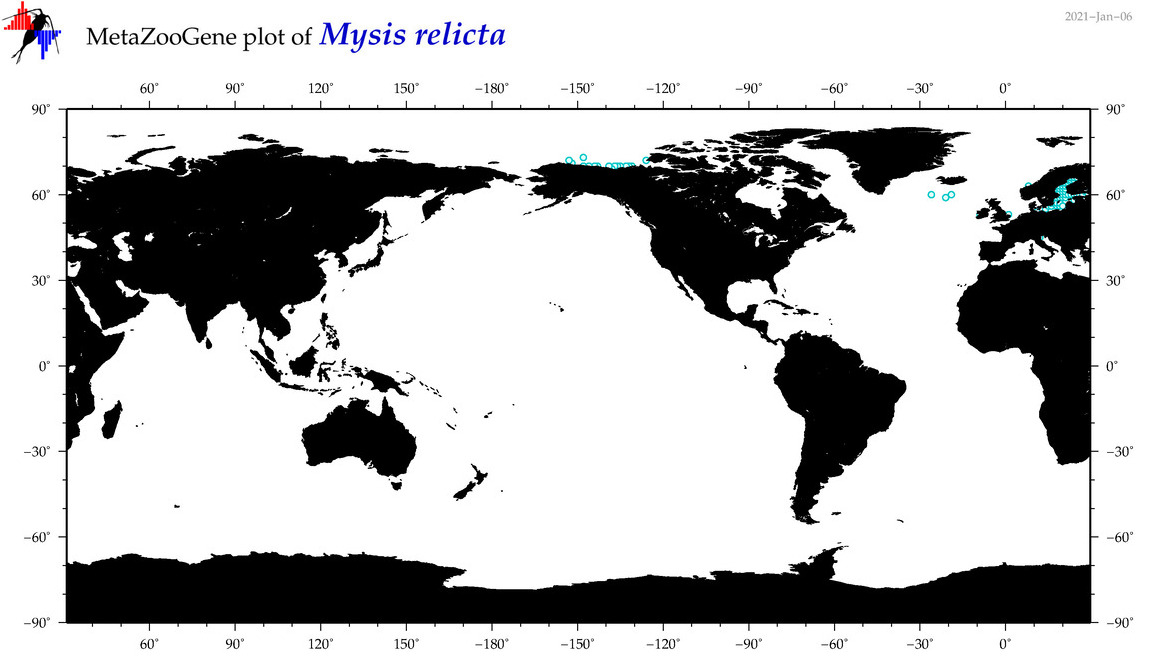 |
accepted
T4001014
R:1:1:1:0 |
| Neomysis integer |
Species
(49) |
COI
12S
16S
18S
28S
|
COI = 48
12S = 0
16S = 1
18S = 5
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
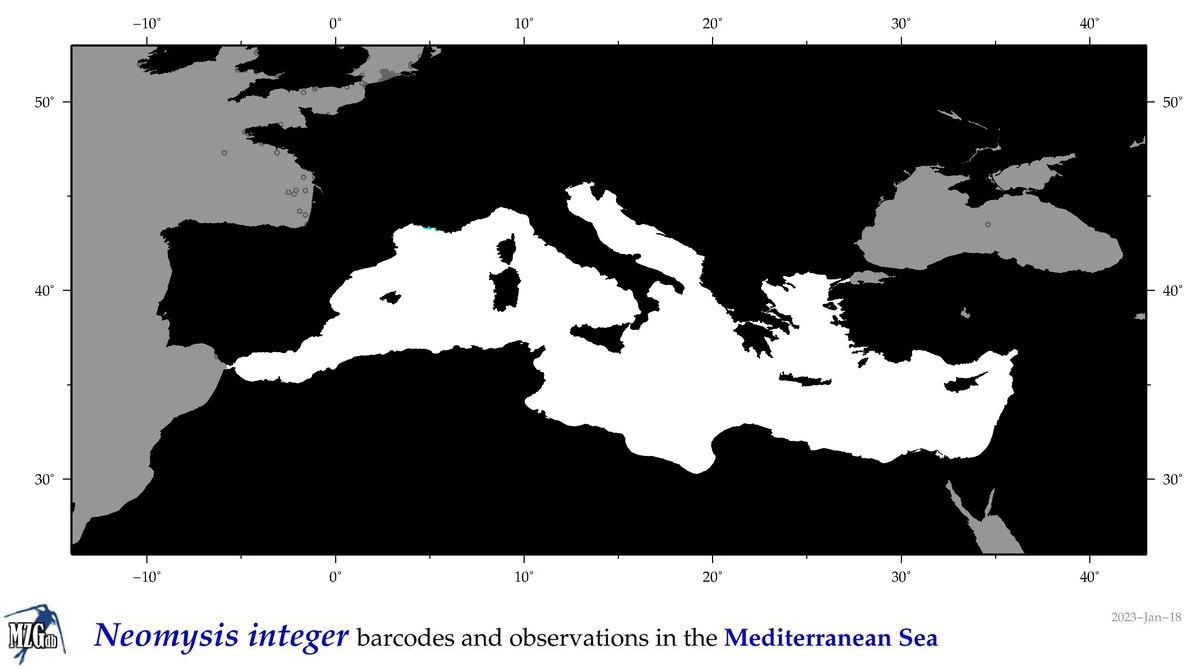 |
accepted
T4003746
R:1:1:0:0 |
| Paraleptomysis apiops |
Species
(50) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
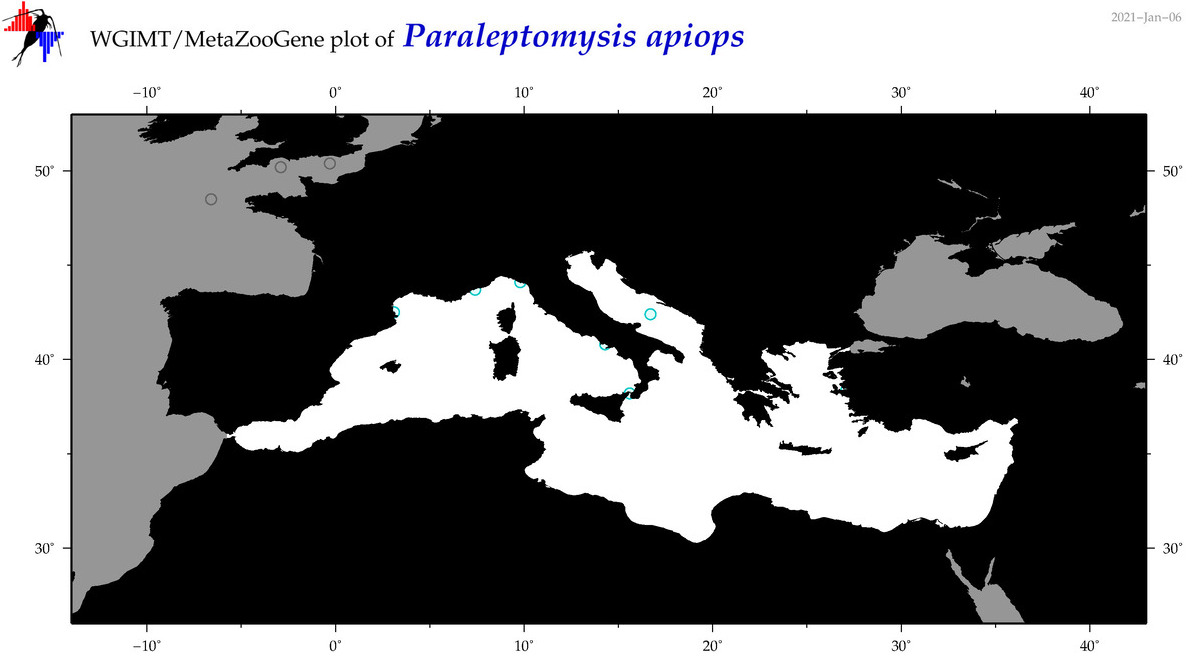 |
accepted
T4062281
R:1:0:0:0 |
| Paraleptomysis banyulensis |
Species
(51) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
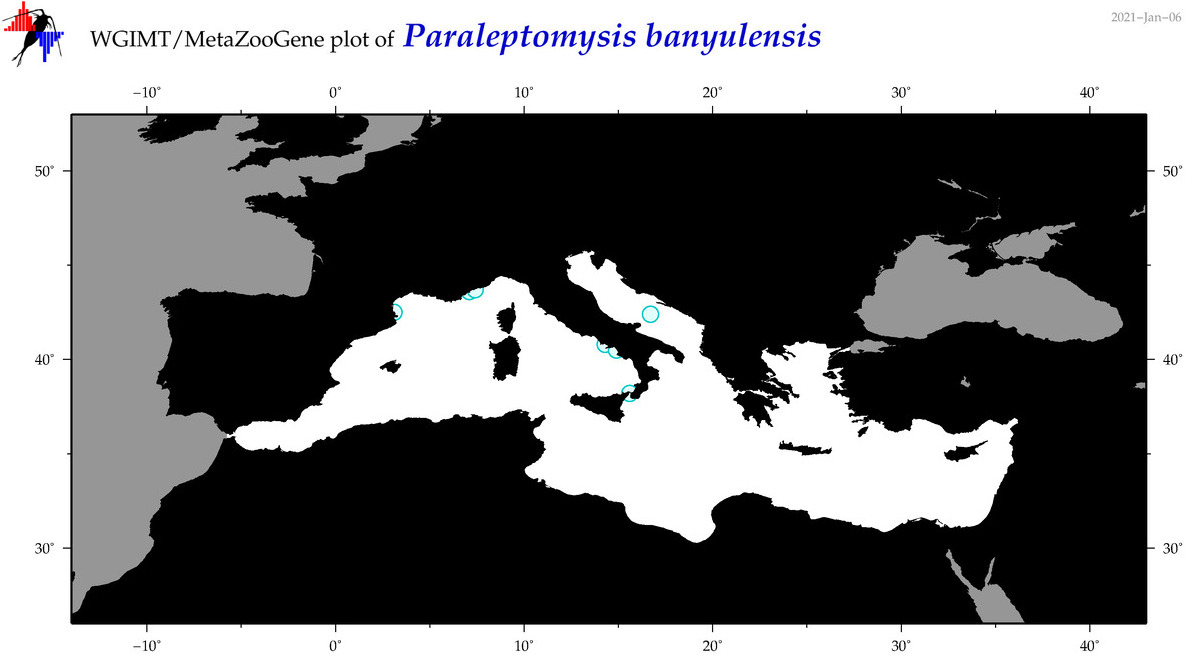 |
accepted
T4062282
R:1:0:0:0 |
| Paramblyops rostratus |
Species
(52) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4062360
R:1:0:0:0 |
| Paramysis arenosa |
Species
(53) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 0
16S = 0
18S = 2
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4007181
R:1:0:0:0 |
| Paramysis (Longidentia) helleri |
Species
(54) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
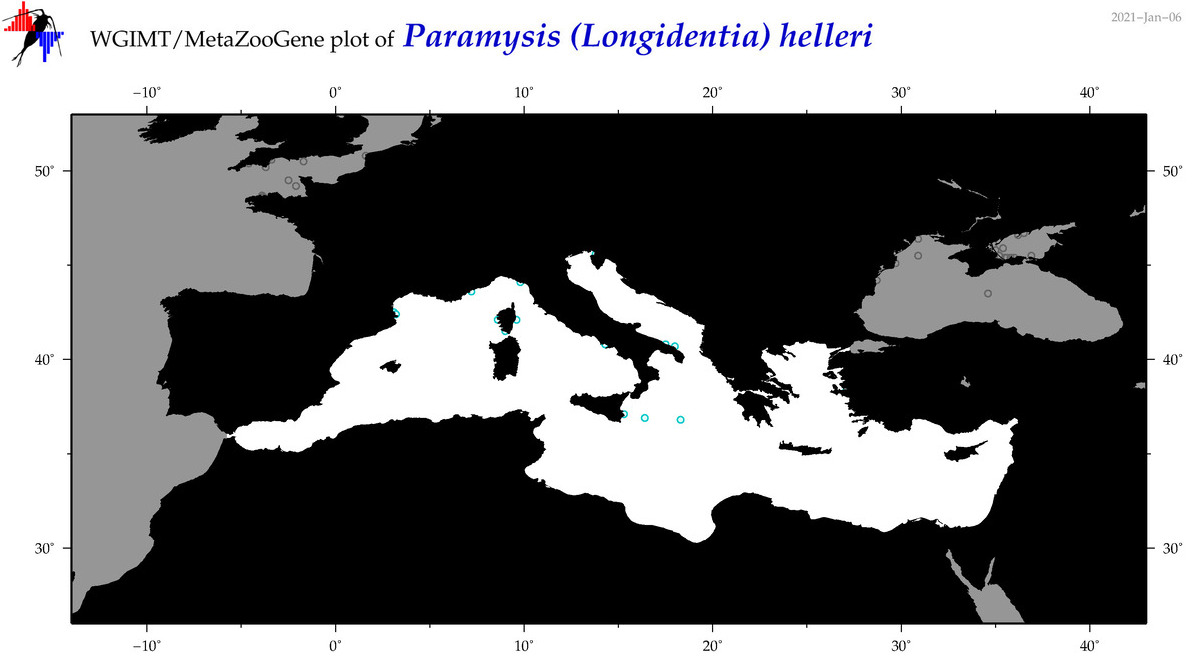 |
accepted
T4007218
R:1:0:1:0 |
| Parapseudomma calloplura |
Species
(55) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4062716
R:1:0:0:0 |
| Parerythrops lobiancoi |
Species
(56) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4062975
R:1:0:0:0 |
| Parerythrops obesus |
Species
(57) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
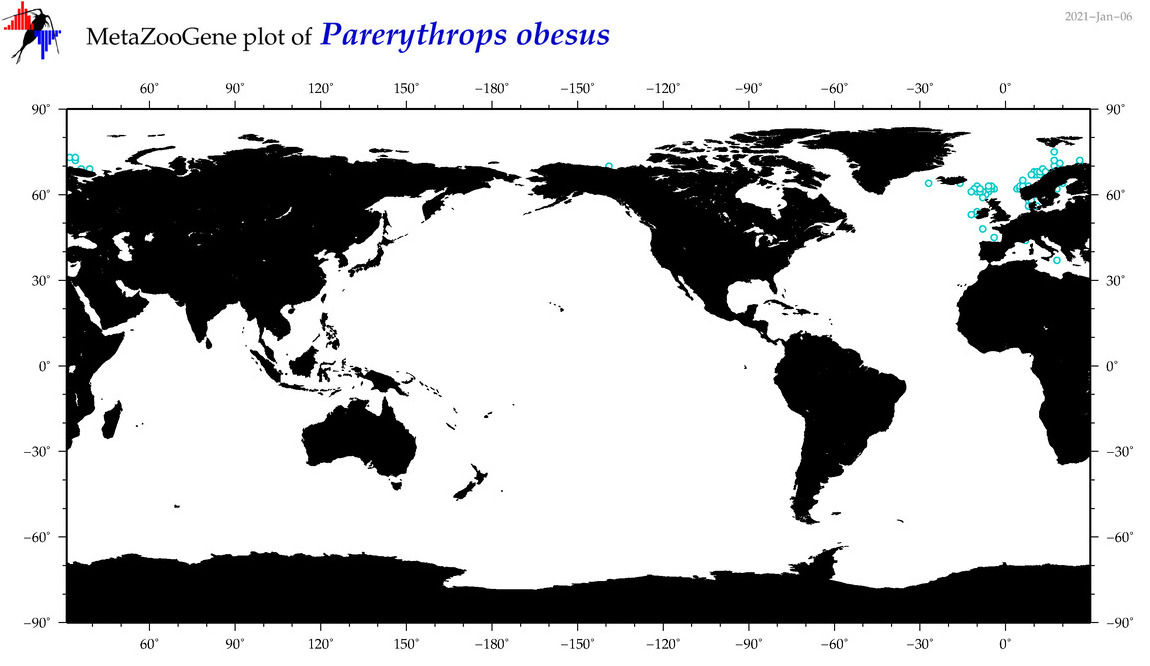 |
accepted
T4001228
R:1:0:0:0 |
| Praunus flexuosus |
Species
(58) |
COI
12S
16S
18S
28S
|
COI = 41
12S = 0
16S = 1
18S = 5
28S = 2
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
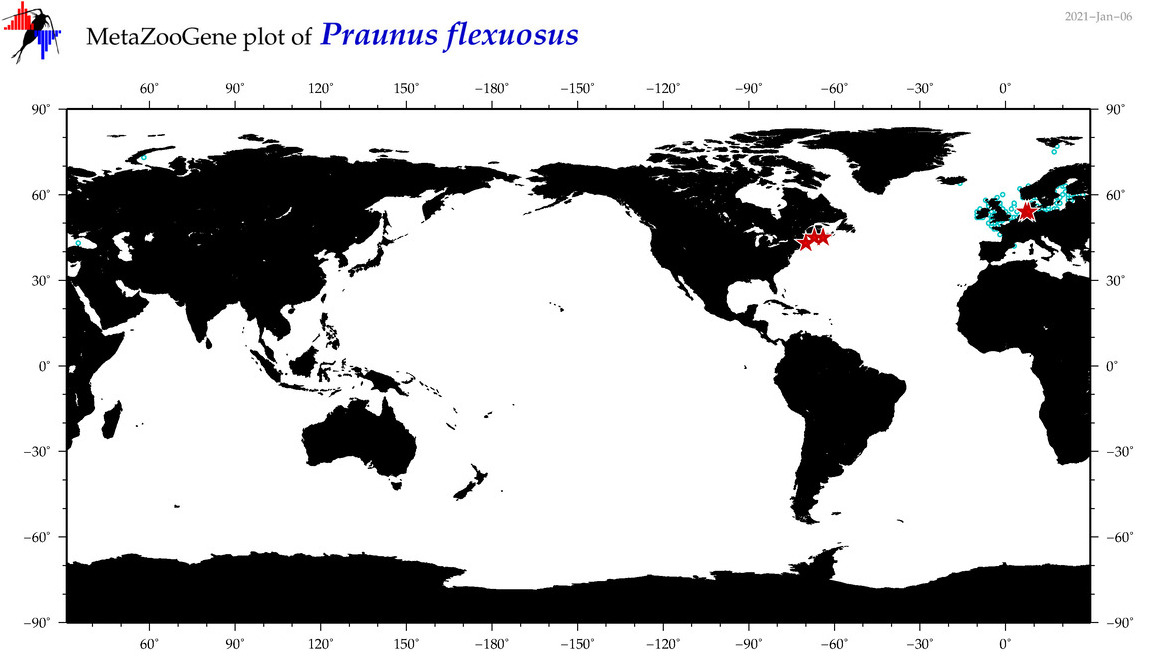 |
accepted
T4007185
R:1:1:0:0 |
| Pseudomma kruppi |
Species
(59) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
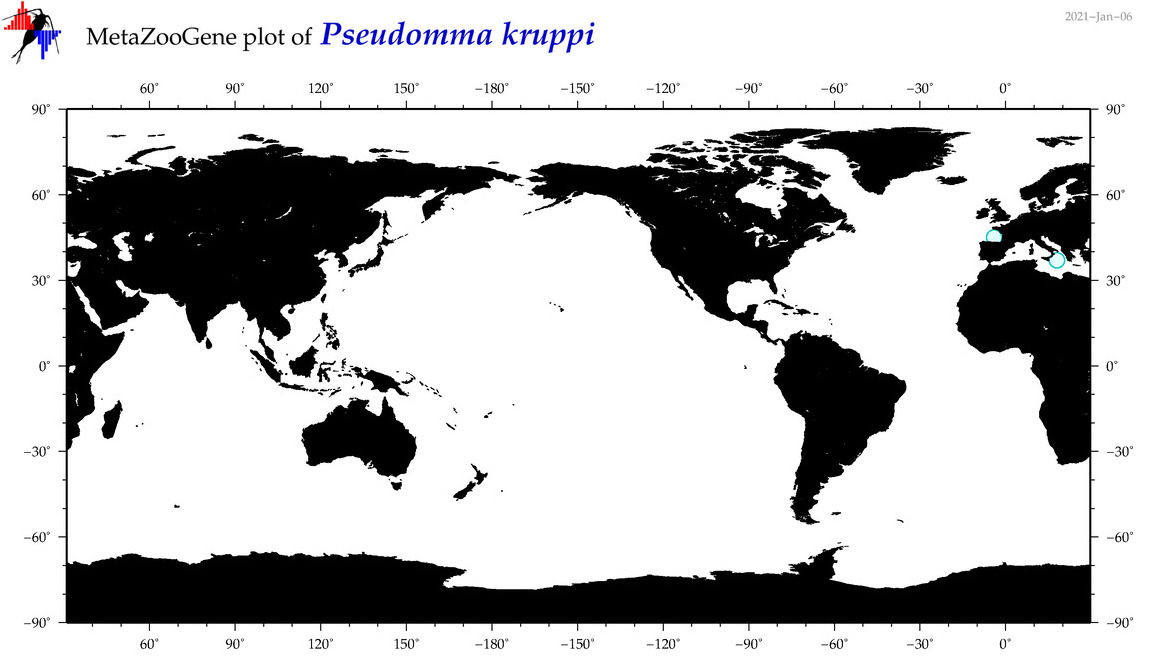 |
accepted
T4014584
R:1:0:0:0 |
| Pyroleptomysis rubra |
Species
(60) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T4066427
R:1:0:0:0 |
| Schistomysis assimilis |
Species
(61) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 2
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
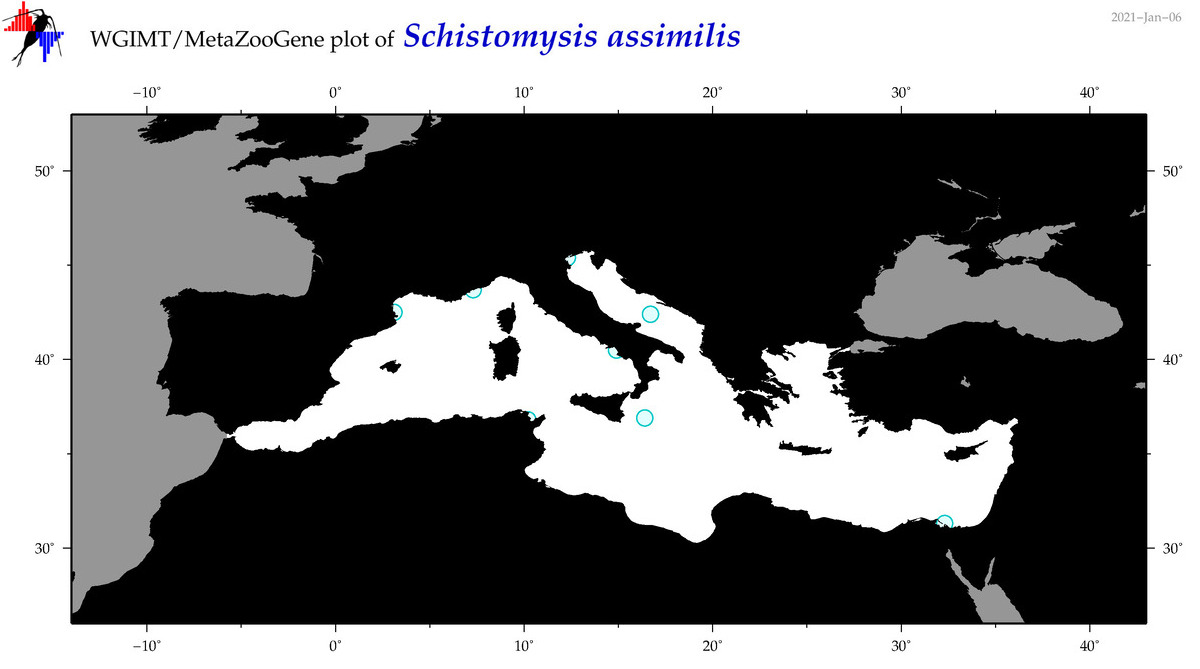 |
accepted
T4014787
R:1:0:0:0 |
| Schistomysis spiritus |
Species
(62) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
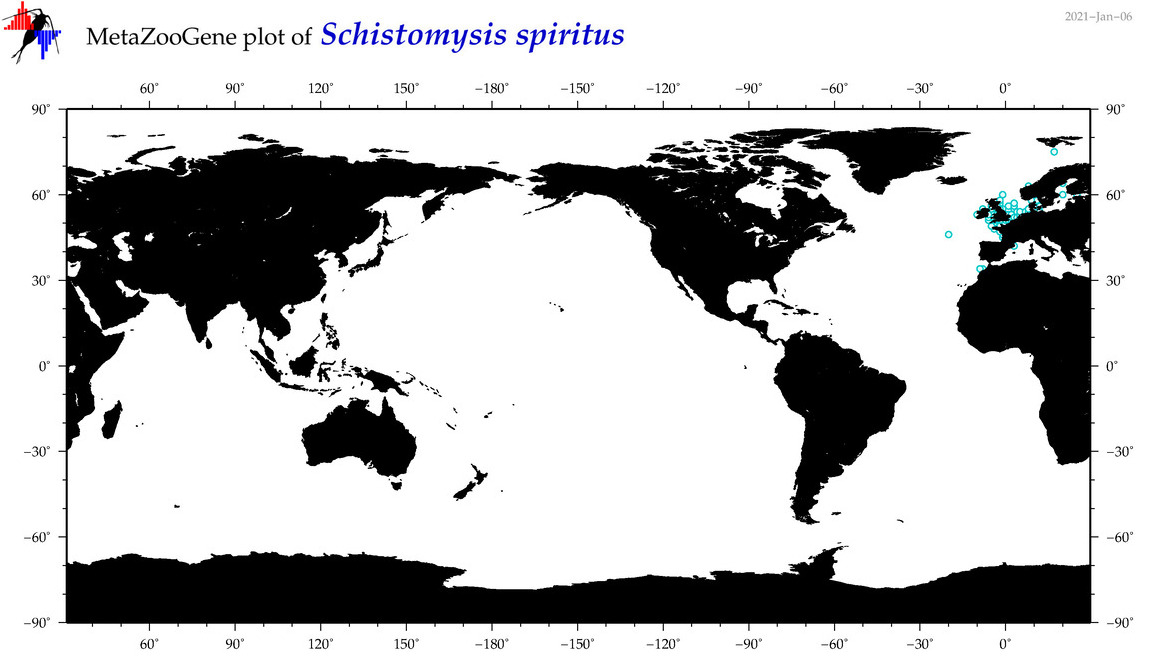 |
accepted
T4007200
R:1:0:0:0 |
| Siriella armata |
Species
(63) |
COI
12S
16S
18S
28S
|
COI = 6
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
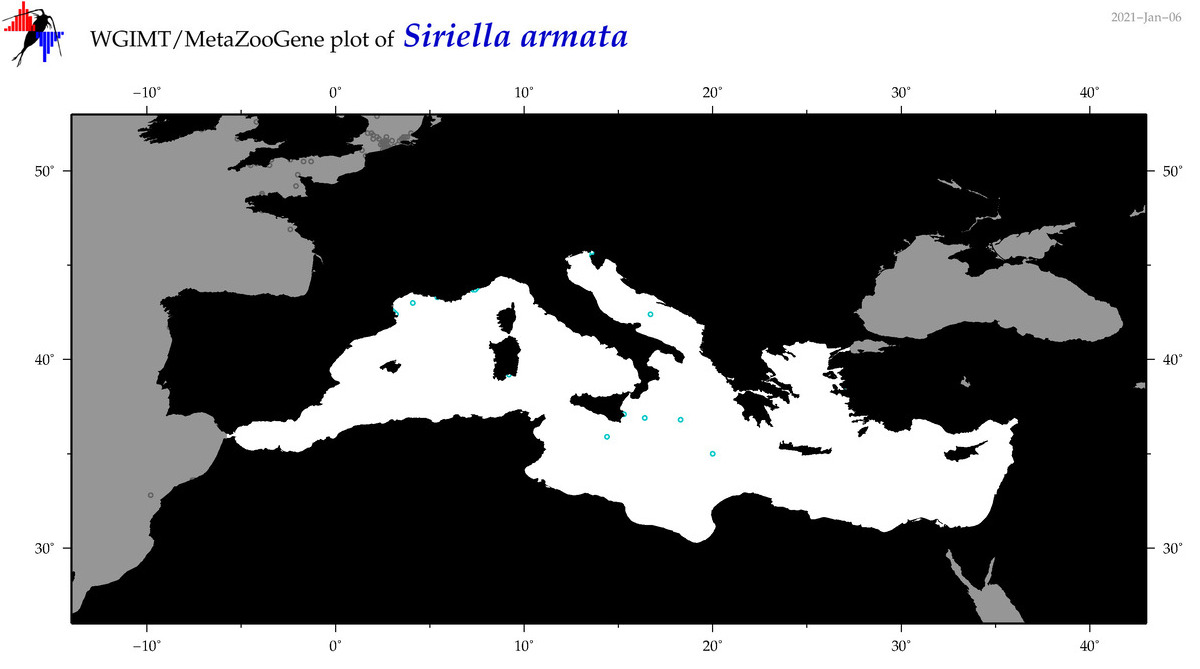 |
accepted
T4007202
R:1:0:0:0 |
| Siriella clausii |
Species
(64) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
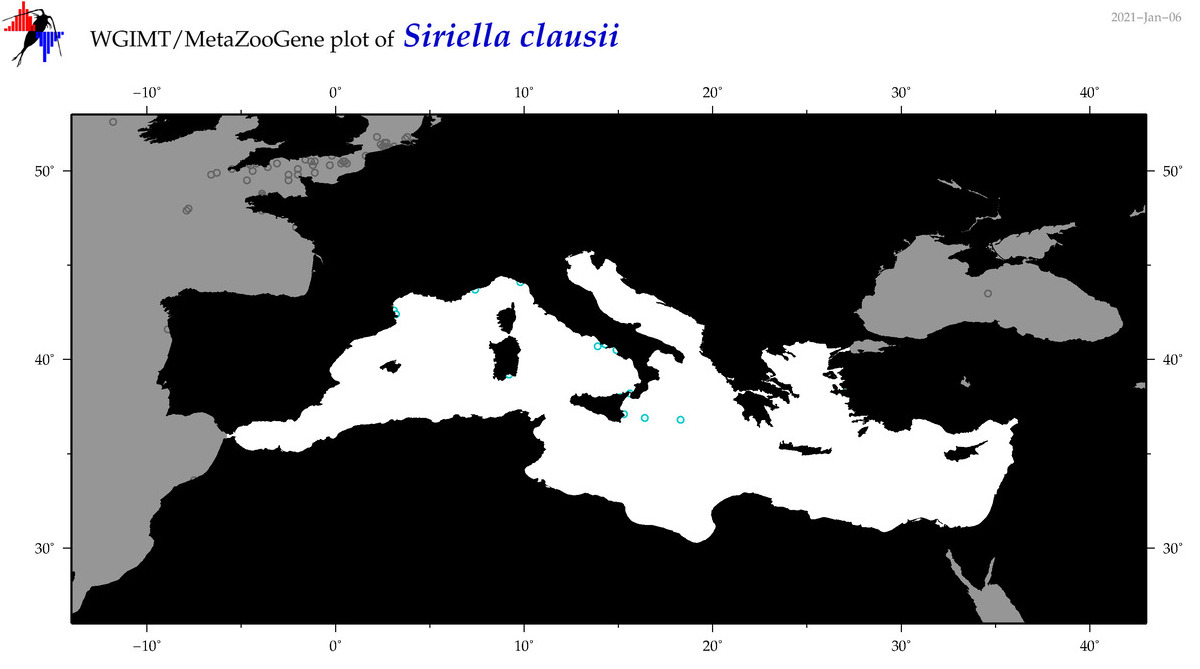 |
accepted
T4007203
R:1:0:0:0 |
| Siriella gracilipes |
Species
(65) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4014973
R:1:0:0:0 |
| Siriella jaltensis |
Species
(66) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 0
16S = 0
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
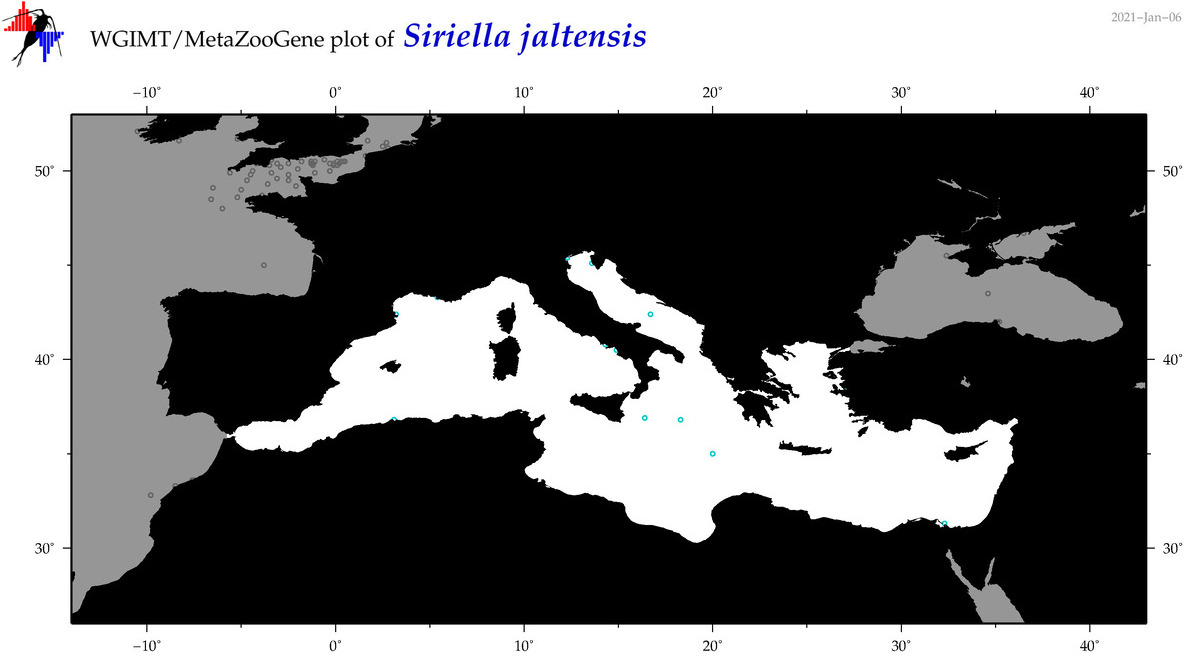 |
accepted
T4007206
R:1:0:0:0 |
| Siriella norvegica |
Species
(67) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
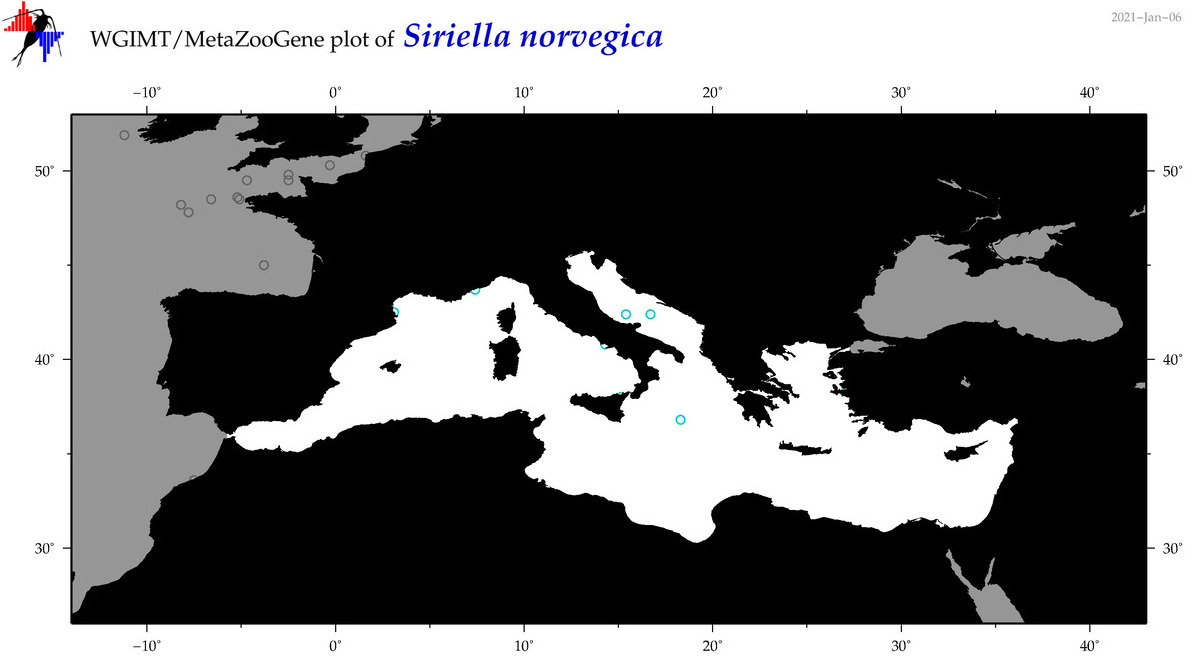 |
accepted
T4007207
R:1:0:0:0 |
| Siriella thompsonii |
Species
(68) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T4007208
R:1:0:0:0 |