Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-26 |
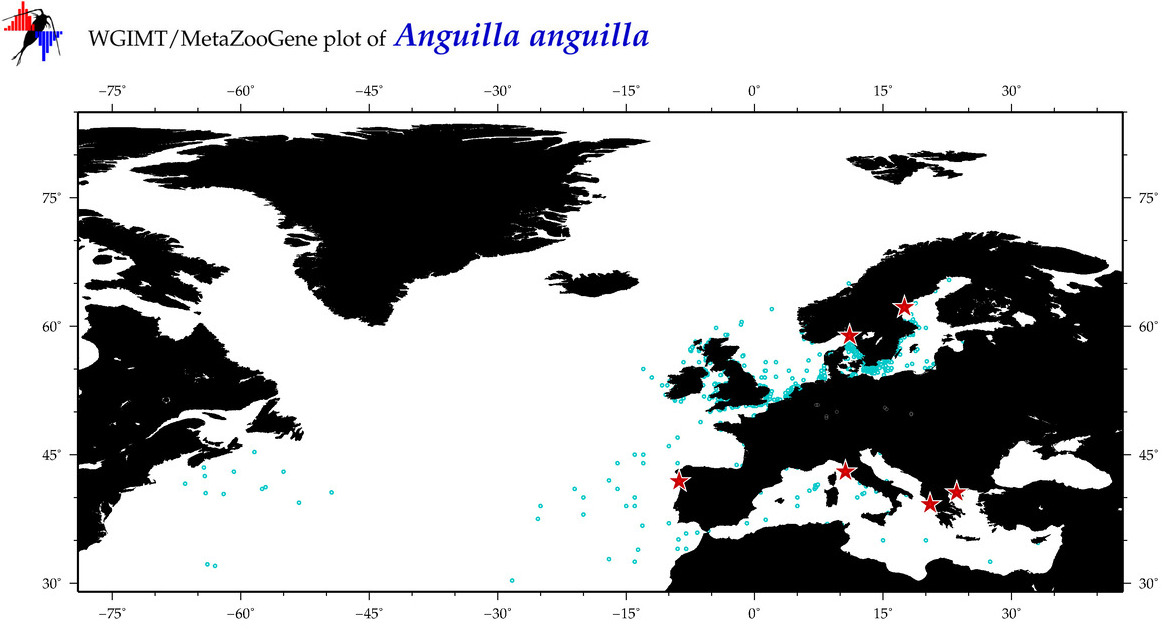
| Anguilla anguilla |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 276
12S = 63
16S = 94
18S = 11
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003105
R:1:1:1:0 |
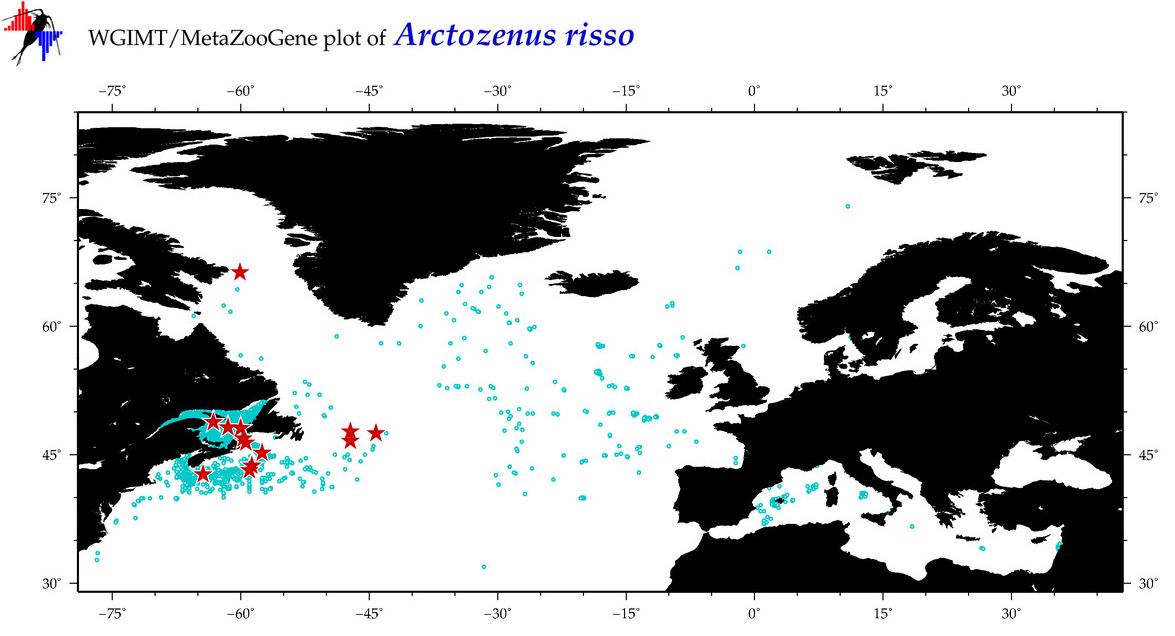
| Arctozenus risso |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 37
12S = 4
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000192
R:1:0:0:0 |
| Argyropelecus hemigymnus |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 52
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000204
R:1:0:0:0 |
| Benthosema glaciale |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 92
12S = 3
16S = 11
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000282
R:1:0:0:0 |
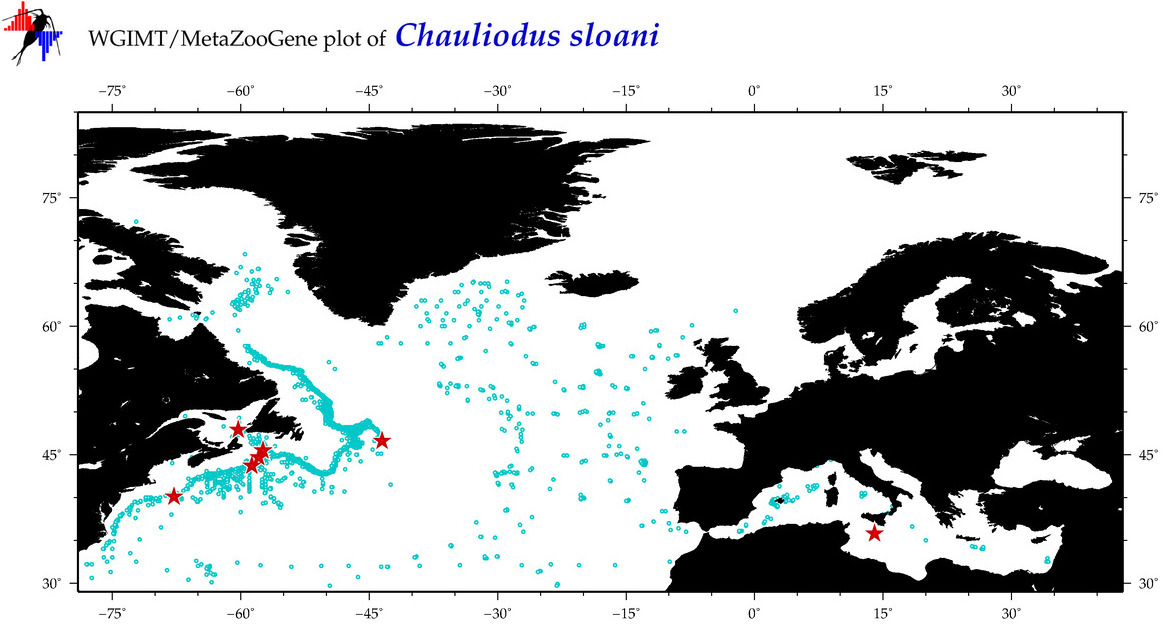
| Chauliodus sloani |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 141
12S = 8
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000382
R:1:0:0:0 |
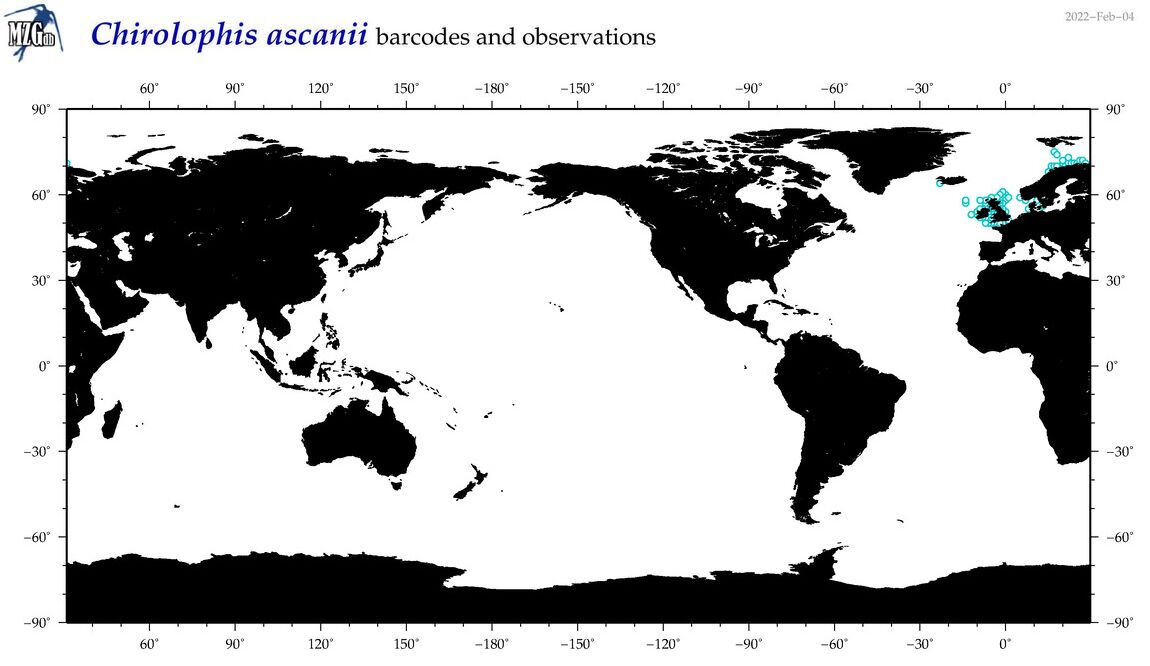
| Chirolophis ascanii |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5018037
R:1:0:0:0 |
| Clupea harengus |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 174
12S = 112
16S = 125
18S = 6
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
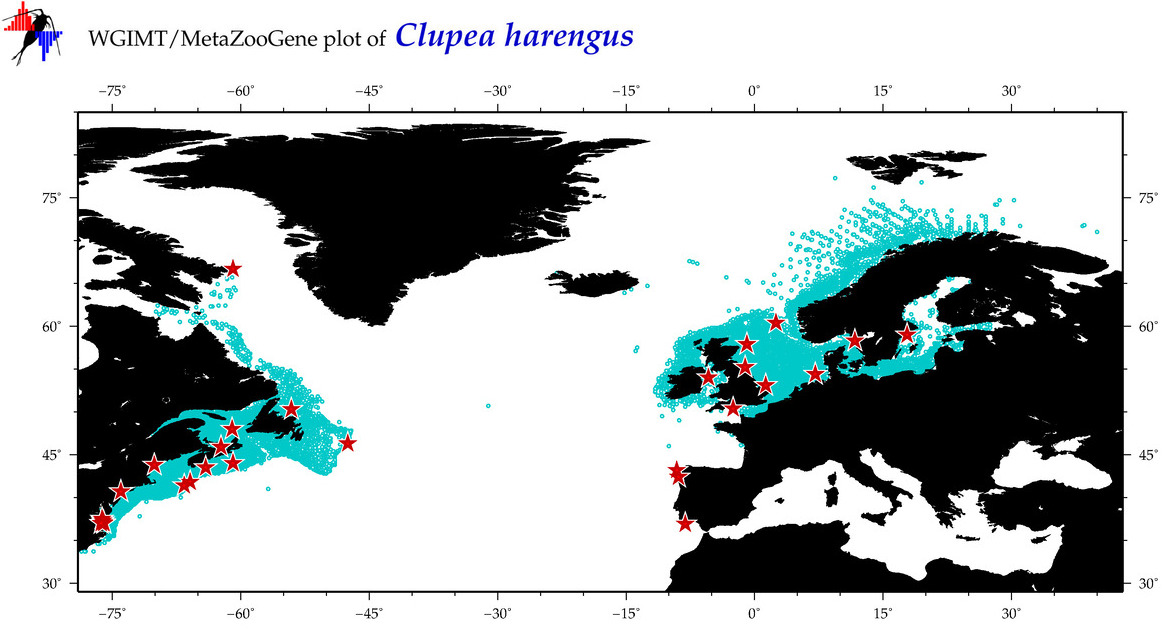 |
accepted
T5000424
R:1:1:1:0 |
| Coryphaenoides rupestris |
Species
(8) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 4
16S = 8
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
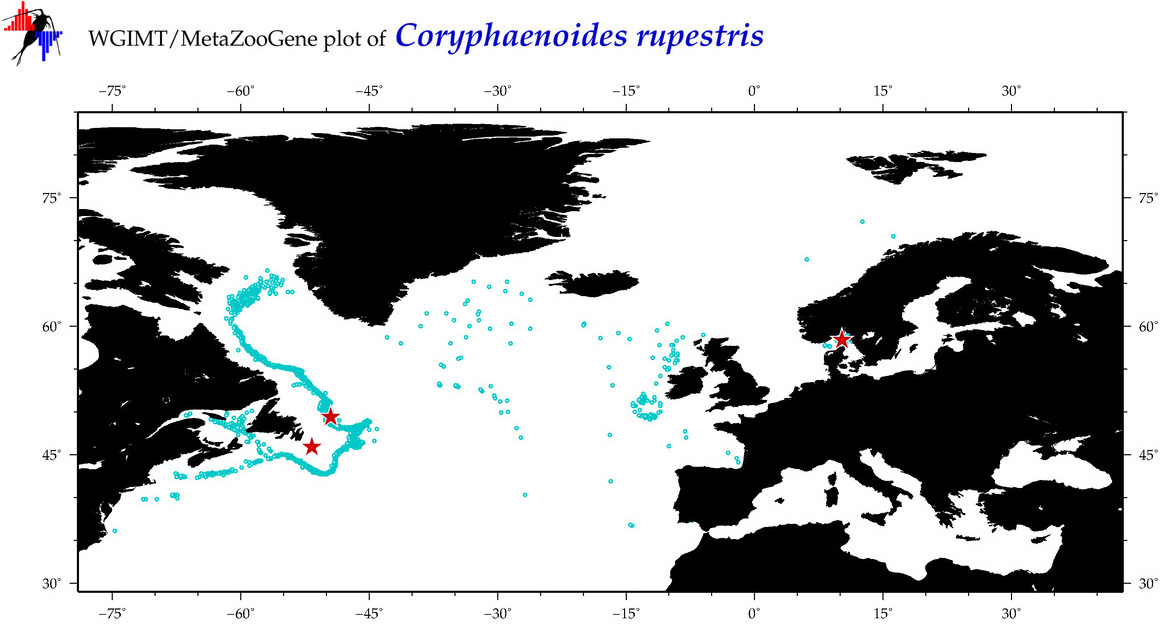 |
accepted
T5003639
R:1:0:0:0 |
| Cottunculus microps |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
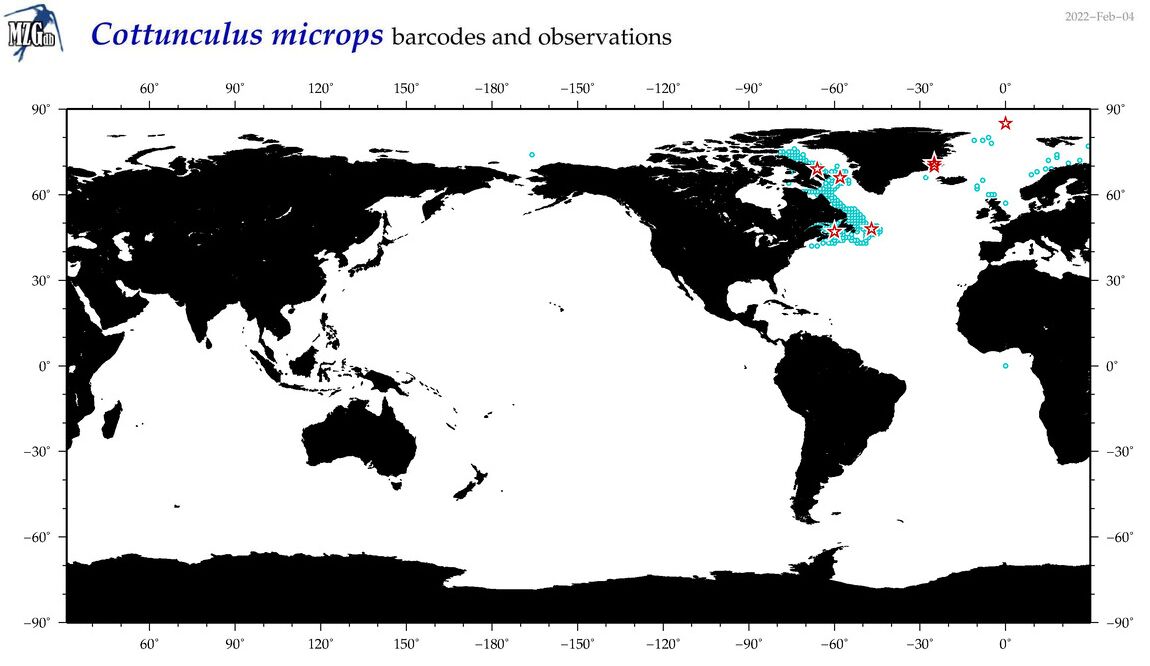 |
accepted
T5003656
R:1:0:0:0 |
| Cyclopterus lumpus |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 67
12S = 6
16S = 9
18S = 0
28S = 2
|
COI = 2
12S = 2
16S = 2
18S = 0
28S = 0
|
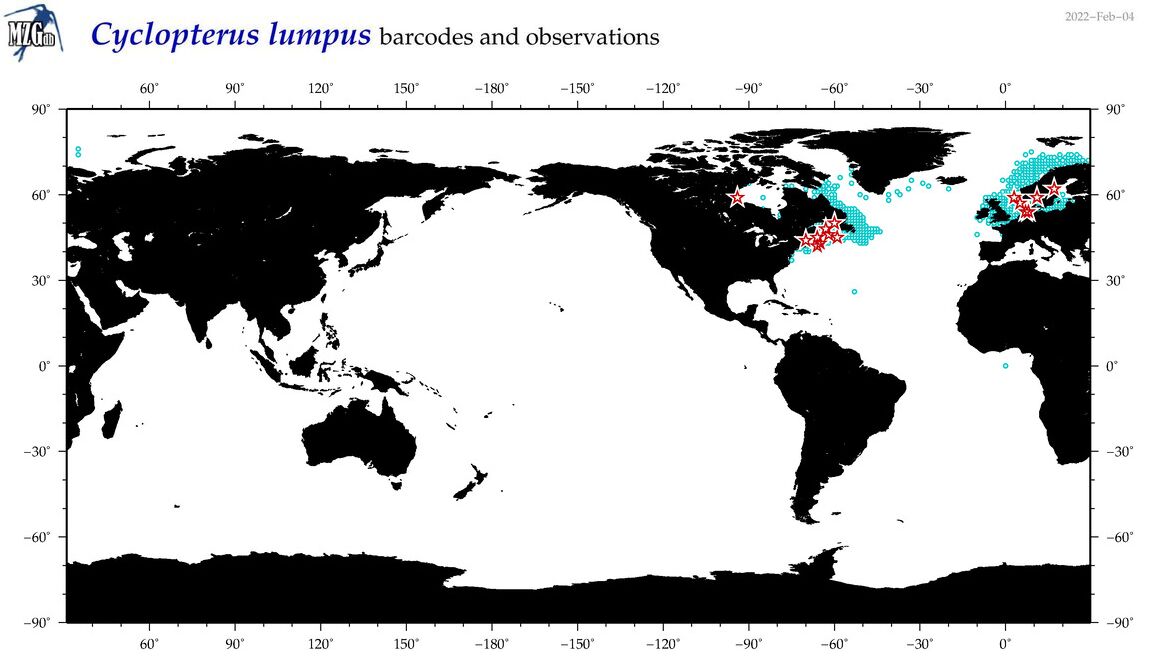 |
accepted
T5000452
R:1:0:0:0 |
| Cyclothone braueri |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 43
12S = 8
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
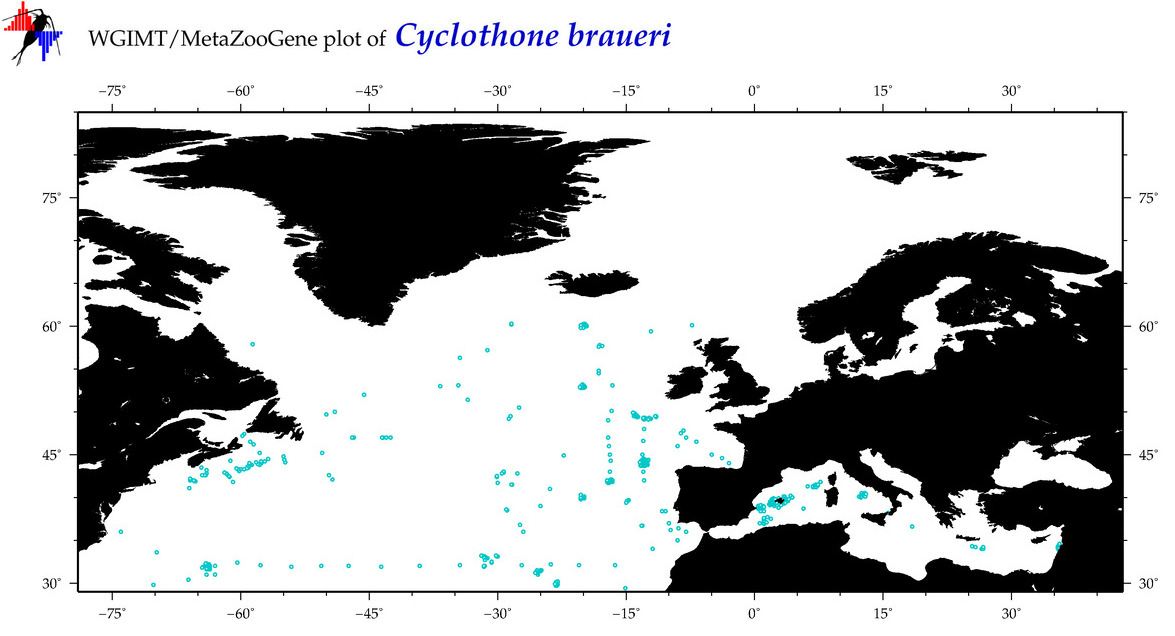 |
accepted
T5000456
R:1:0:0:0 |
| Echiodon drummondii |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5019266
R:1:0:0:0 |
| Enchelyopus cimbrius |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 54
12S = 5
16S = 8
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
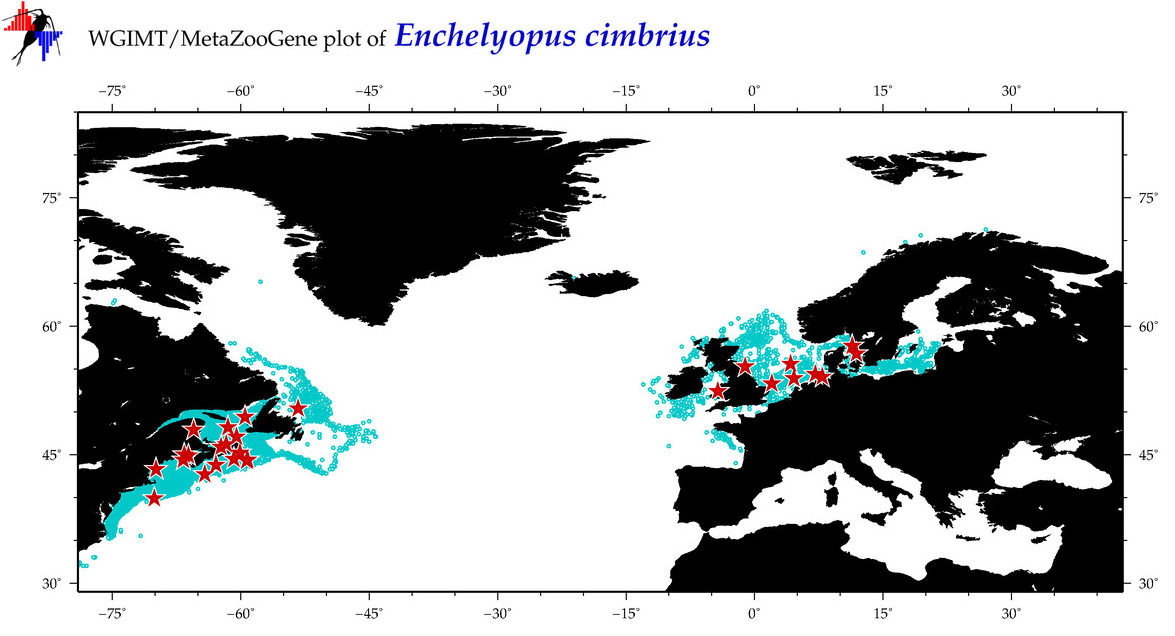 |
accepted
T5000541
R:1:0:0:0 |
| Gadiculus argenteus |
Species
(14) |
COI
12S
16S
18S
28S
|
COI = 37
12S = 4
16S = 6
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004037
R:1:0:0:0 |
| Gadiculus thori |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
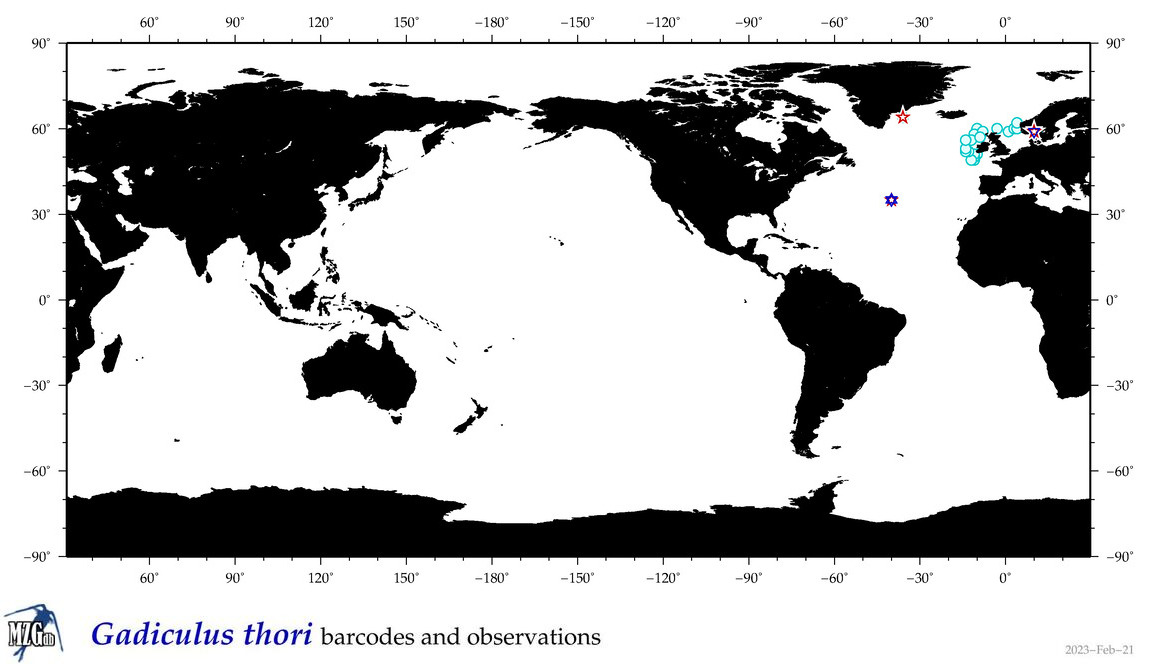 |
accepted
T5019985
R:1:0:0:0 |
| Gadus morhua |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 627
12S = 296
16S = 305
18S = 7
28S = 3
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000588
R:1:1:0:0 |
| Gaidropsarus argentatus |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 31
12S = 4
16S = 3
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
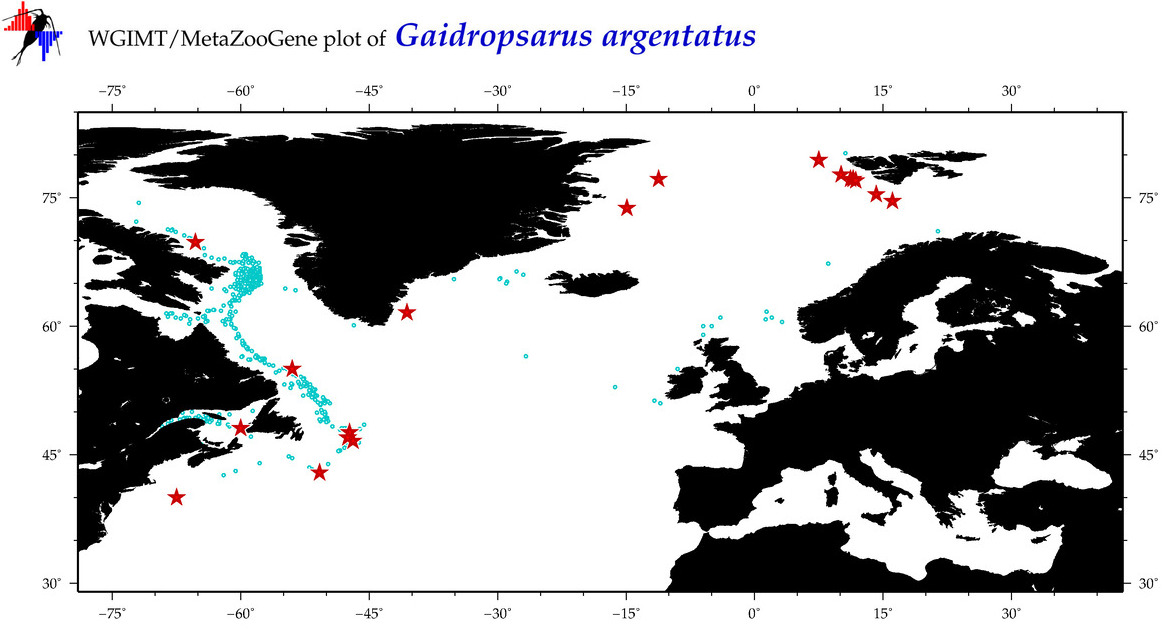 |
accepted
T5004039
R:1:0:0:0 |
| Gaidropsarus ensis |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 38
12S = 6
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
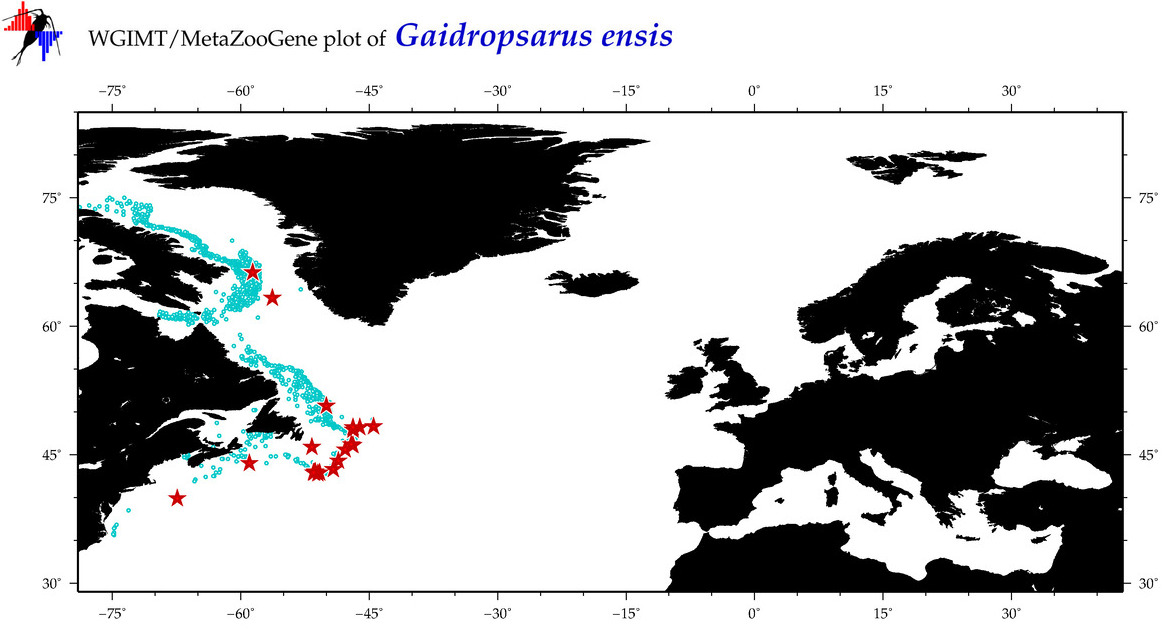 |
accepted
T5004041
R:1:0:0:0 |
| Glyptocephalus cynoglossus |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 57
12S = 2
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
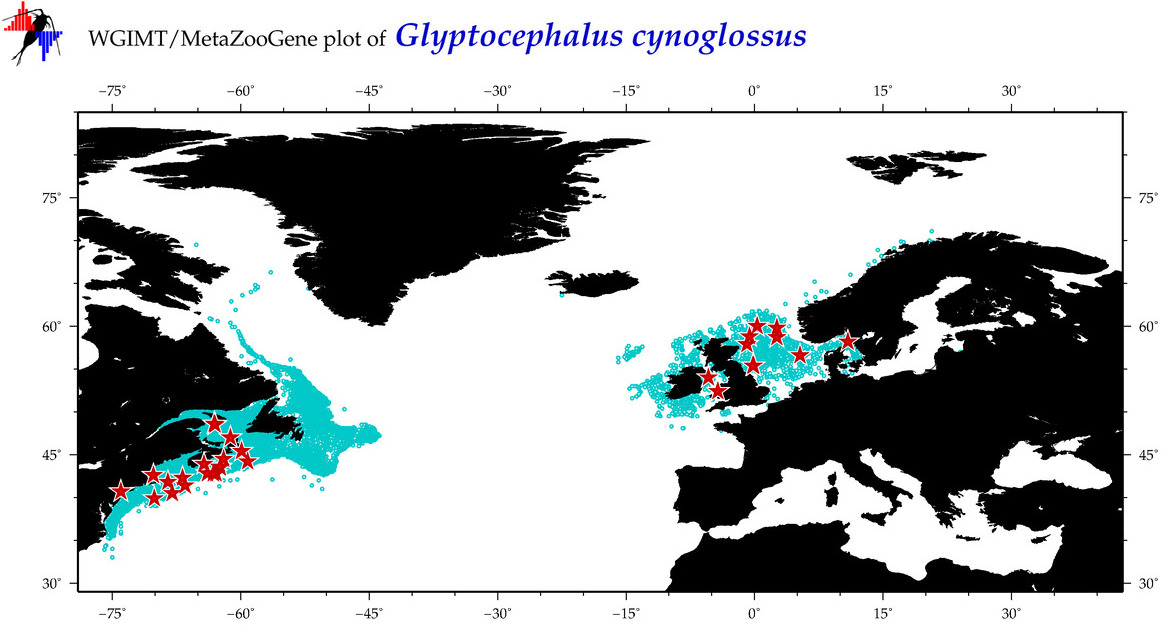 |
accepted
T5000602
R:1:0:0:0 |
| Halargyreus johnsonii |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 89
12S = 2
16S = 7
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004183
R:1:0:0:0 |
| Hippoglossoides platessoides |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 40
12S = 5
16S = 7
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000673
R:1:0:0:0 |
| Icelus bicornis |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
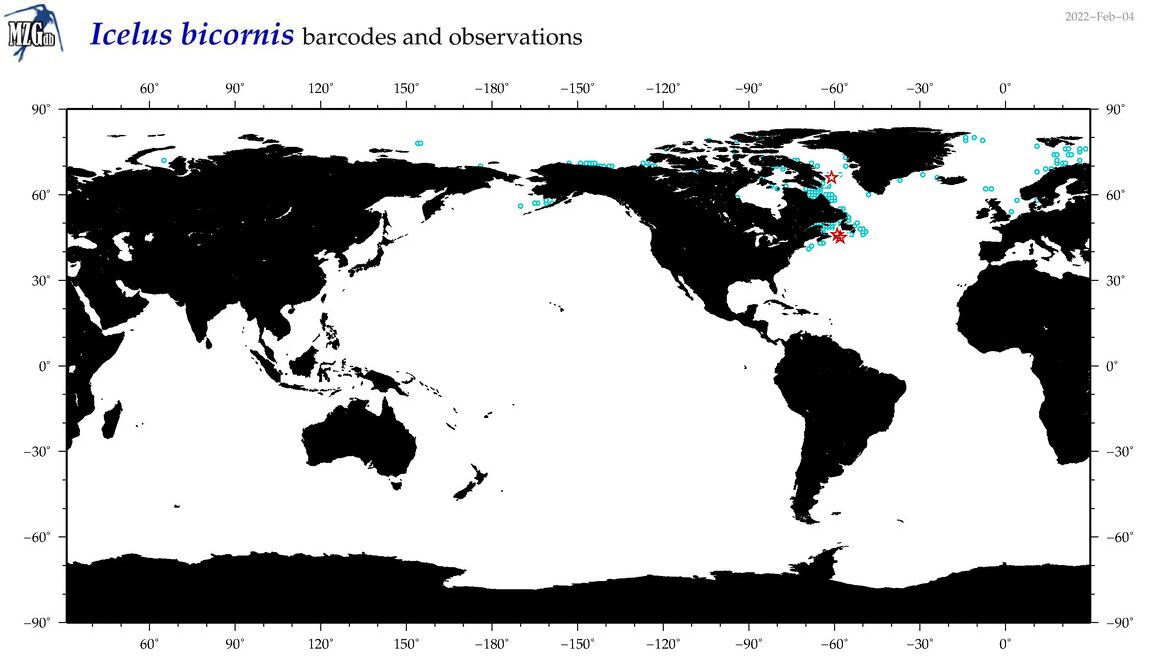 |
accepted
T5004342
R:1:0:0:0 |
| Lampanyctus intricarius |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 4
16S = 6
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
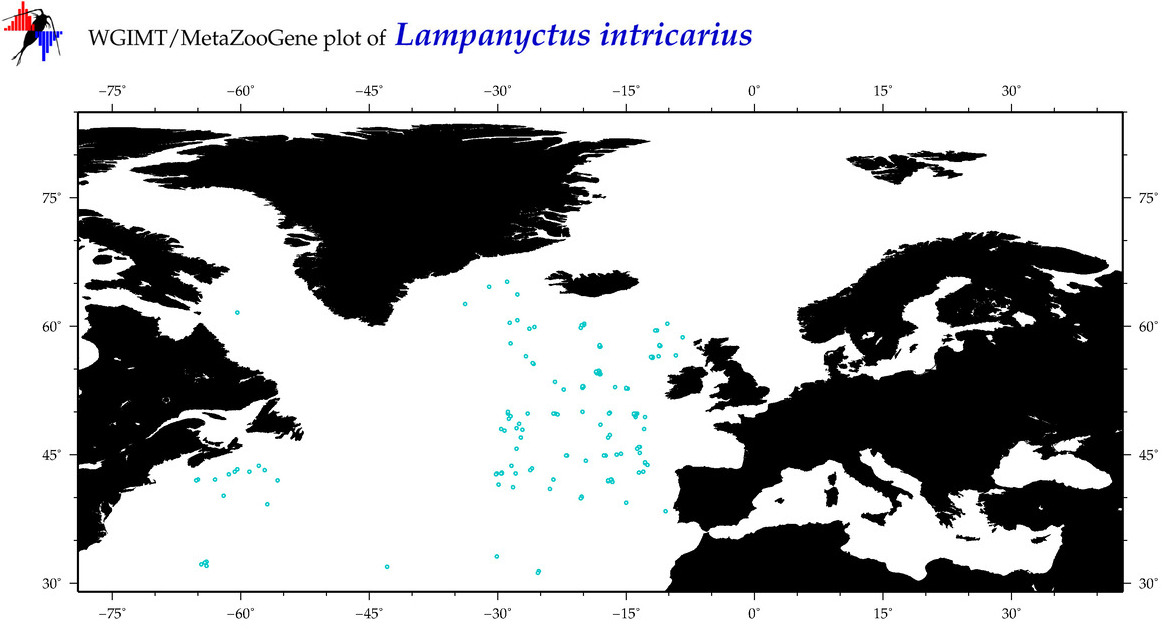 |
accepted
T5004411
R:1:0:0:0 |
| Lepidion eques |
Species
(24) |
COI
12S
16S
18S
28S
|
COI = 32
12S = 1
16S = 16
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004431
R:1:0:0:0 |
| Lycodes squamiventer |
Species
(25) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
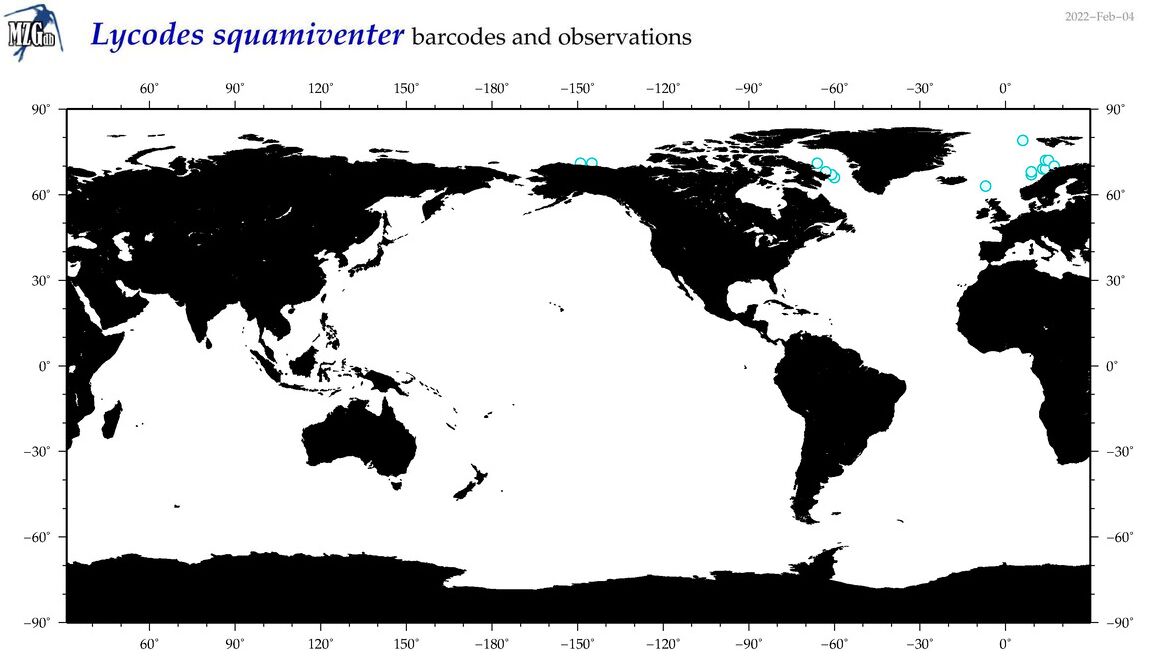 |
accepted
T5021905
R:1:0:0:0 |
| Maurolicus muelleri |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 122
12S = 4
16S = 18
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000872
R:1:0:0:0 |
| Melanogrammus aeglefinus |
Species
(27) |
COI
12S
16S
18S
28S
|
COI = 176
12S = 10
16S = 16
18S = 4
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
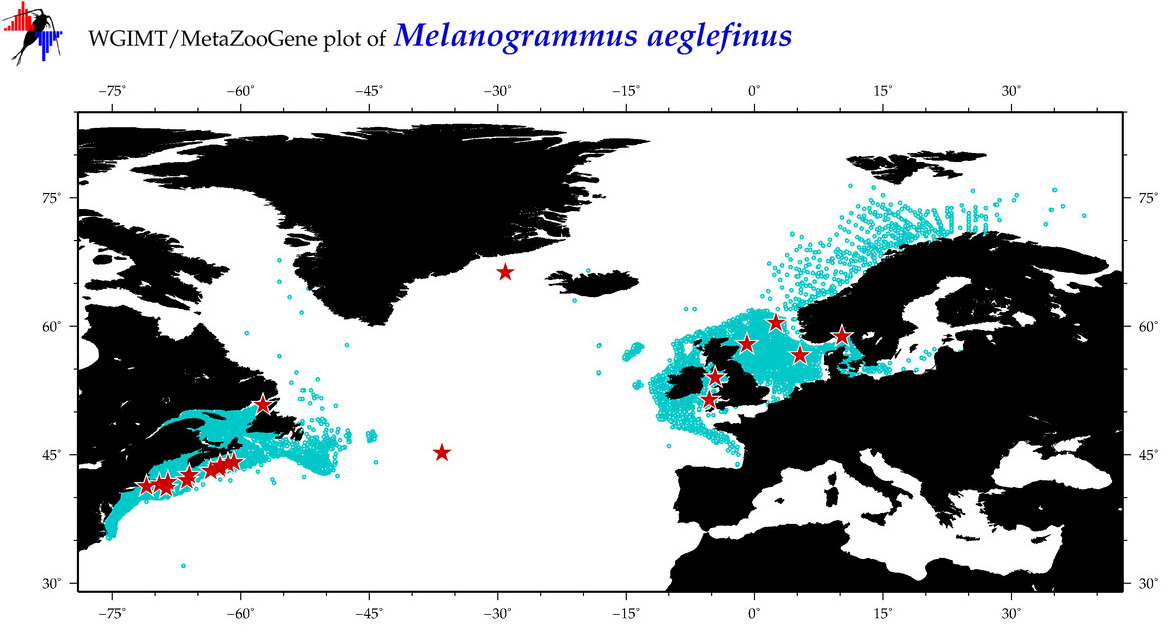 |
accepted
T5000884
R:1:0:0:0 |
| Merlangius merlangus |
Species
(28) |
COI
12S
16S
18S
28S
|
COI = 111
12S = 7
16S = 13
18S = 3
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004652
R:1:1:0:0 |
| Micromesistius poutassou |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 122
12S = 9
16S = 8
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
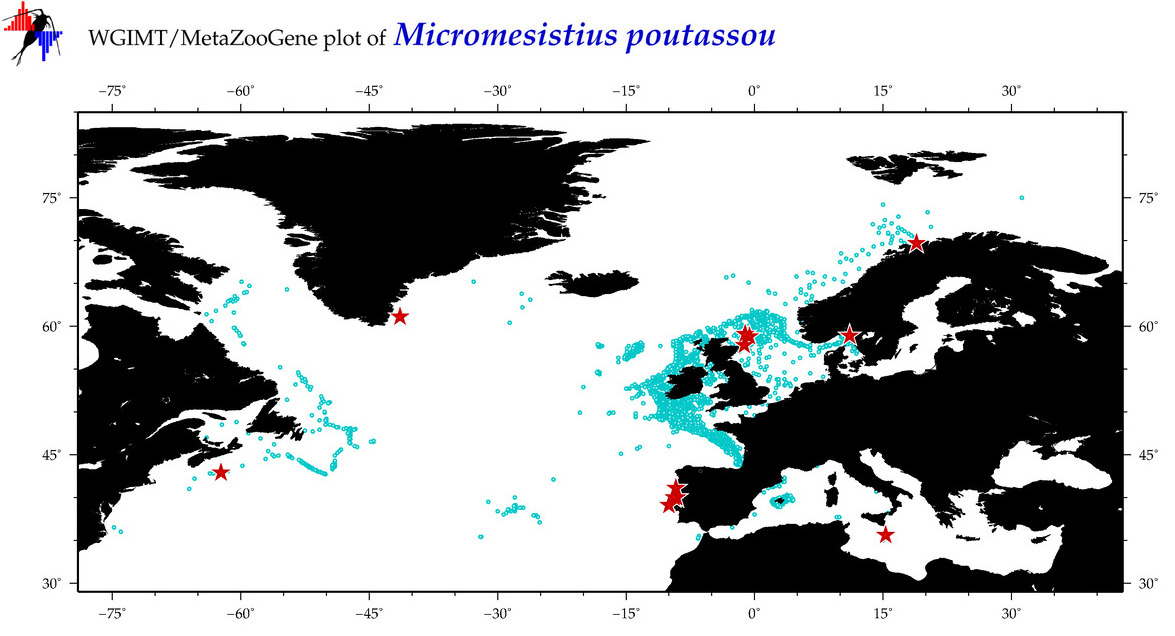 |
accepted
T5004677
R:1:0:0:0 |
| Molva dypterygia |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 5
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
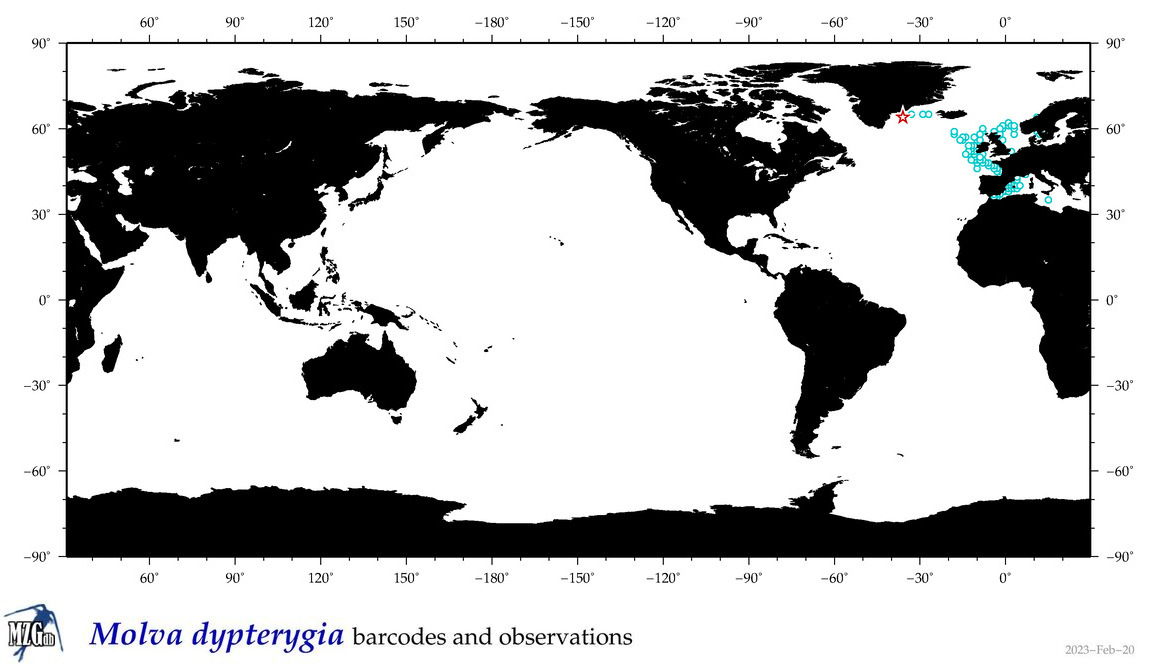 |
accepted
T5022259
R:1:0:0:0 |
| Myoxocephalus scorpius |
Species
(31) |
COI
12S
16S
18S
28S
|
COI = 85
12S = 6
16S = 6
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
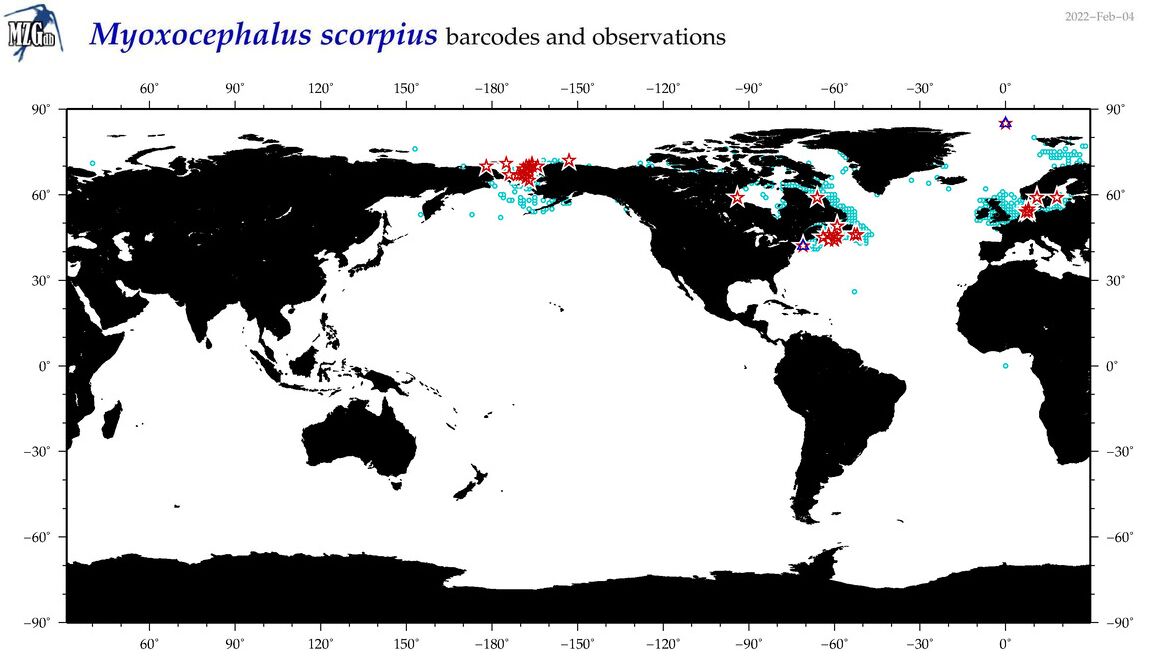 |
accepted
T5000952
R:1:1:0:0 |
| Nemichthys scolopaceus |
Species
(32) |
COI
12S
16S
18S
28S
|
COI = 42
12S = 9
16S = 8
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000976
R:1:0:0:0 |
| Nezumia aequalis |
Species
(33) |
COI
12S
16S
18S
28S
|
COI = 0
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022696
R:1:0:0:0 |
| Notacanthus bonaparte |
Species
(34) |
COI
12S
16S
18S
28S
|
COI = 68
12S = 5
16S = 5
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
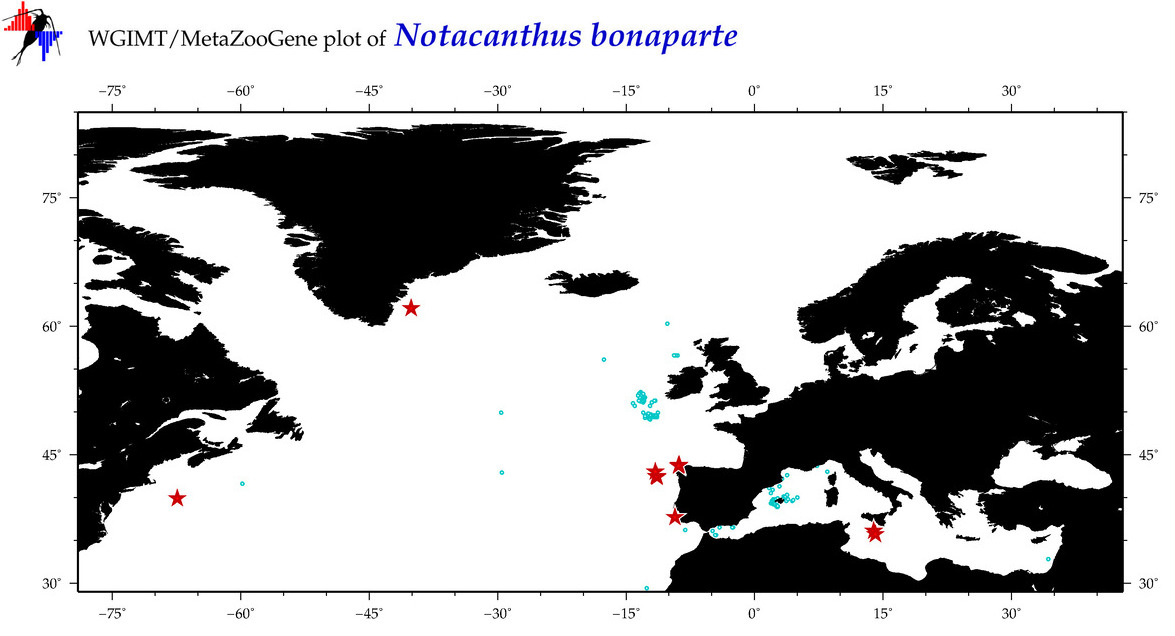 |
accepted
T5004806
R:1:0:0:0 |
| Notoscopelus kroyeri |
Species
(35) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 3
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
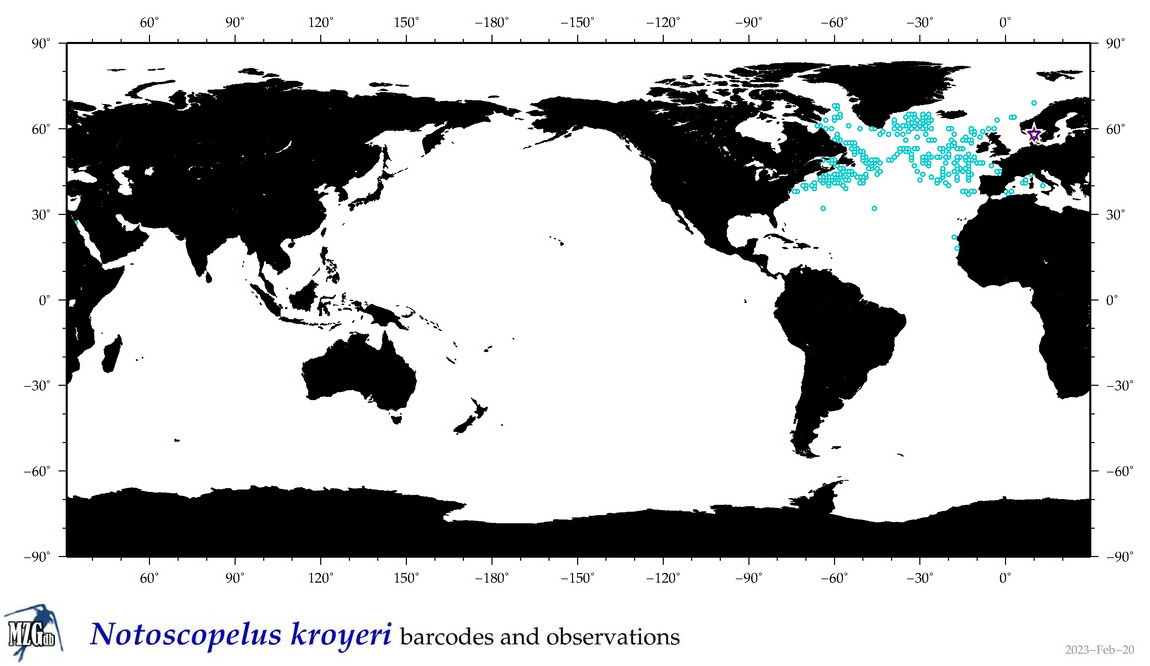 |
accepted
T5022795
R:1:0:0:0 |
| Pollachius virens |
Species
(36) |
COI
12S
16S
18S
28S
|
COI = 137
12S = 8
16S = 8
18S = 1
28S = 4
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
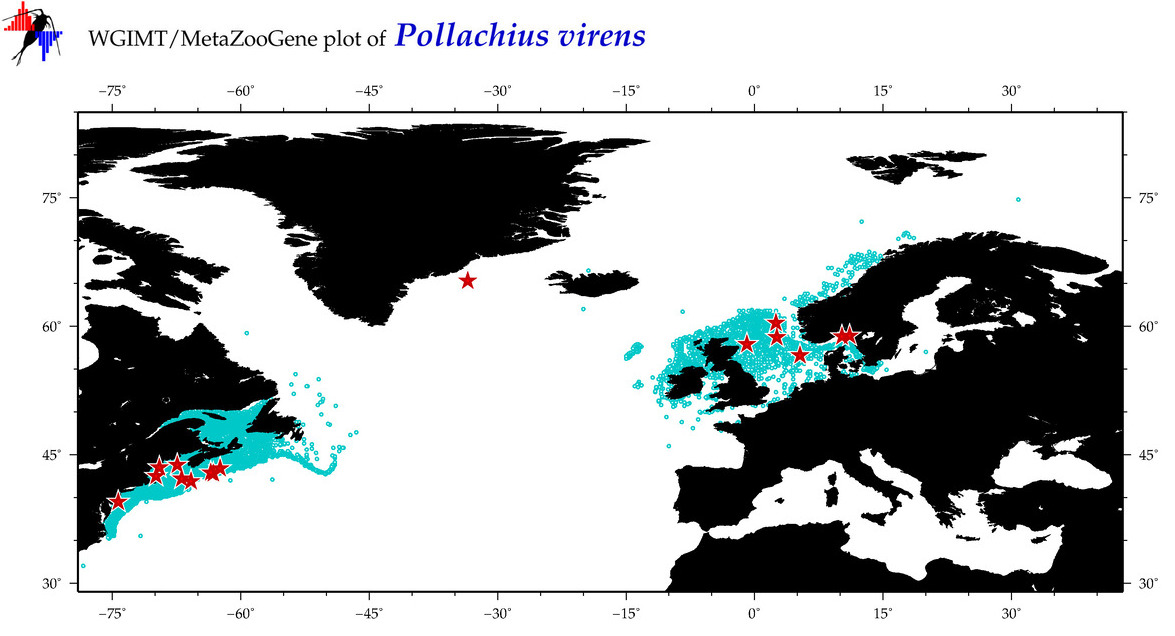 |
accepted
T5001101
R:1:0:0:0 |
| Protomyctophum arcticum |
Species
(37) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 2
16S = 5
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000035
R:1:0:0:0 |
| Salvelinus alpinus |
Species
(38) |
COI
12S
16S
18S
28S
|
COI = 164
12S = 125
16S = 132
18S = 2
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
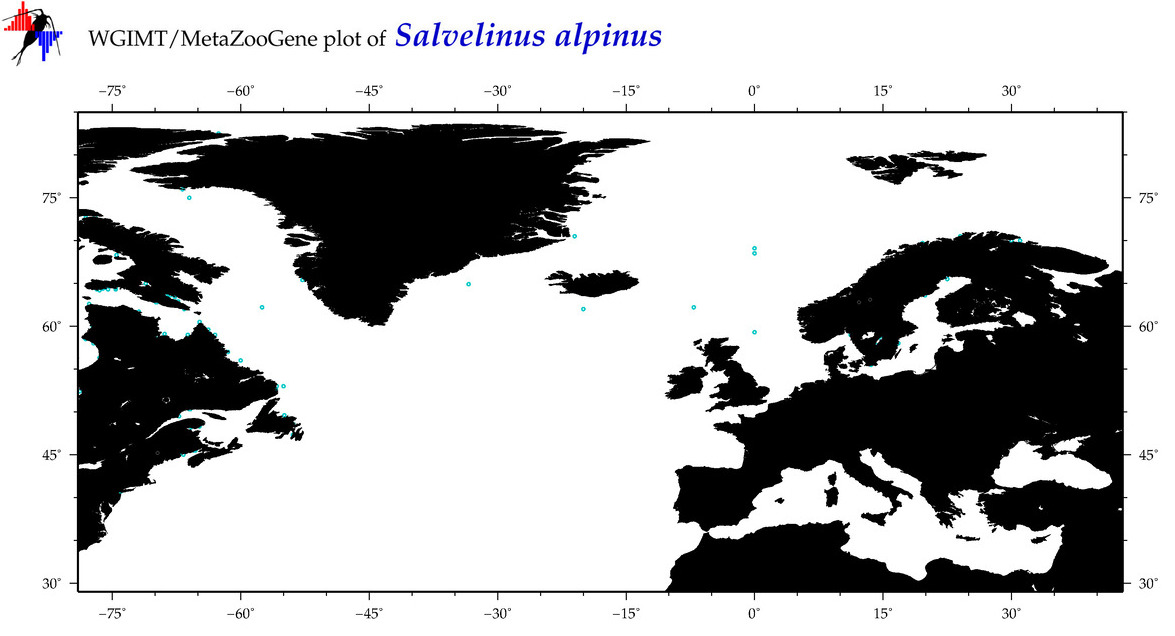 |
accepted
T5012036
R:1:1:1:0 |
| Sebastes mentella |
Species
(39) |
COI
12S
16S
18S
28S
|
COI = 31
12S = 7
16S = 5
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
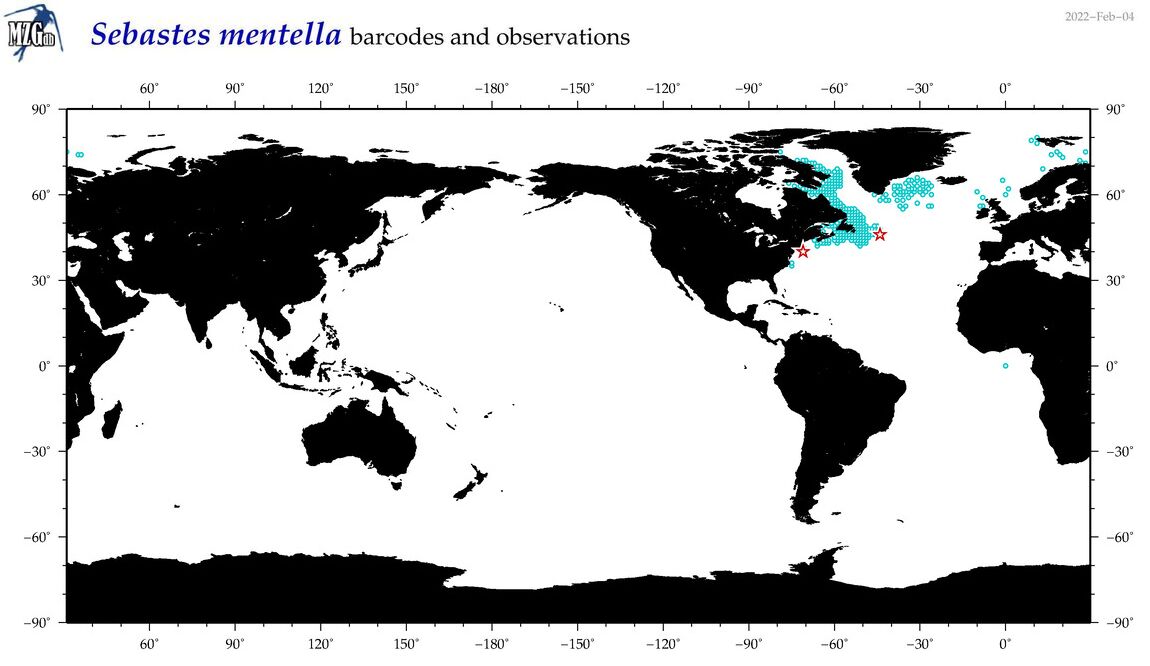 |
accepted
T5005493
R:1:0:0:0 |
| Sebastes viviparus |
Species
(40) |
COI
12S
16S
18S
28S
|
COI = 11
12S = 5
16S = 5
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
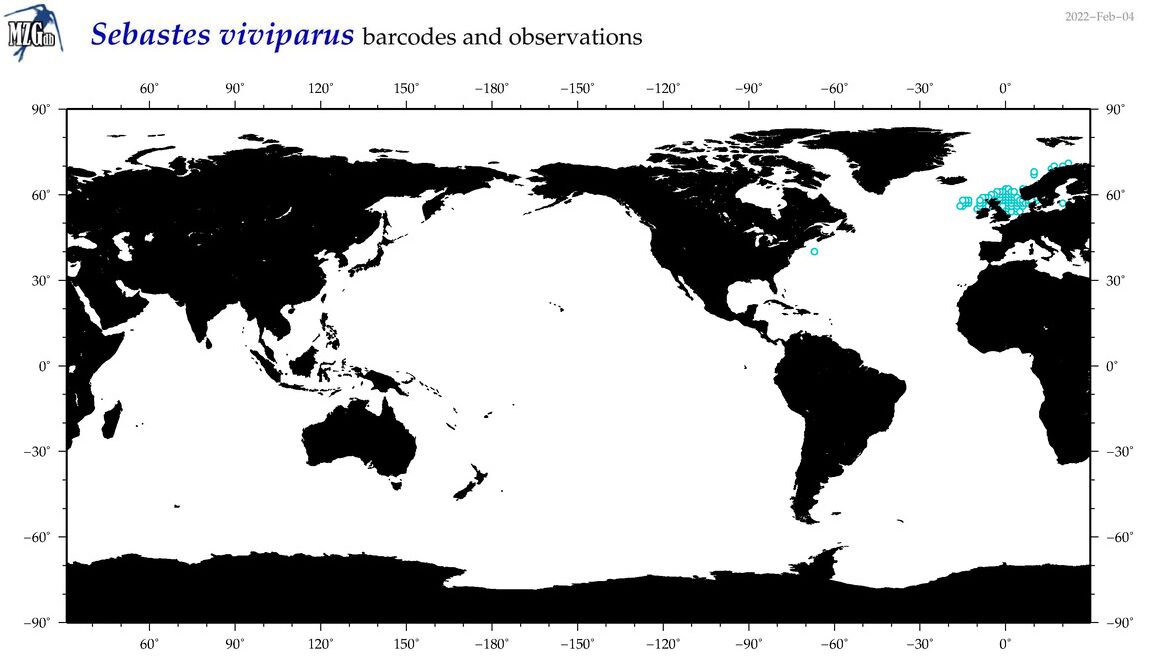 |
accepted
T5005535
R:1:0:0:0 |
| Synaphobranchus kaupii |
Species
(41) |
COI
12S
16S
18S
28S
|
COI = 124
12S = 7
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
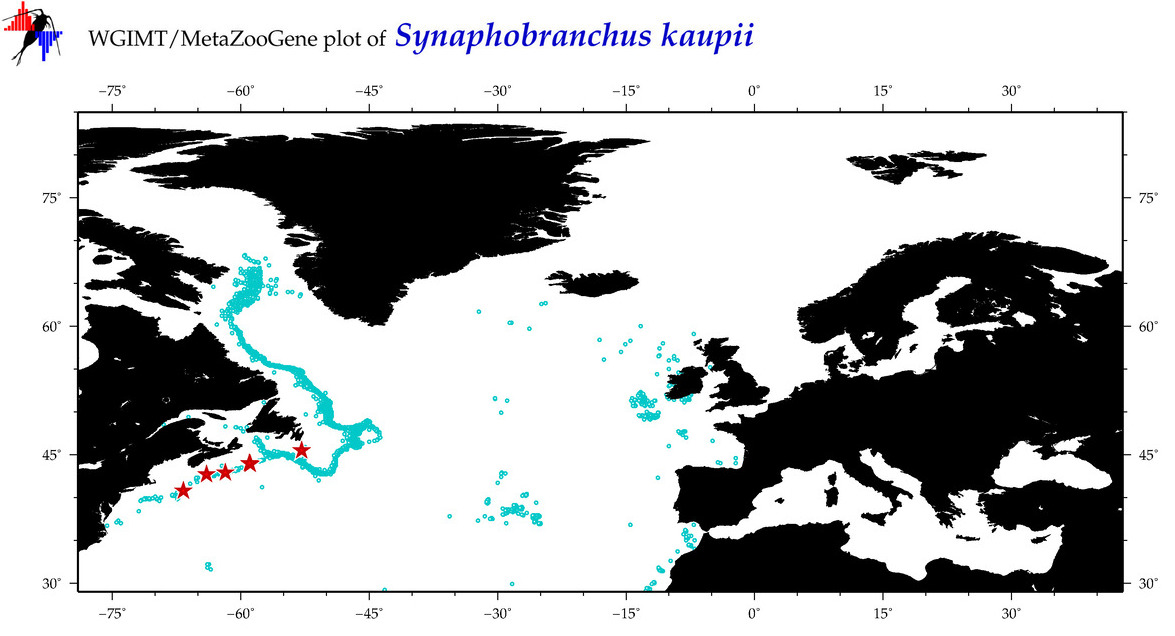 |
accepted
T5012412
R:1:0:0:0 |
| Trachyrincus murrayi |
Species
(42) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 4
16S = 2
18S = 0
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026339
R:1:0:0:0 |
| Triglops murrayi |
Species
(43) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 3
16S = 2
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
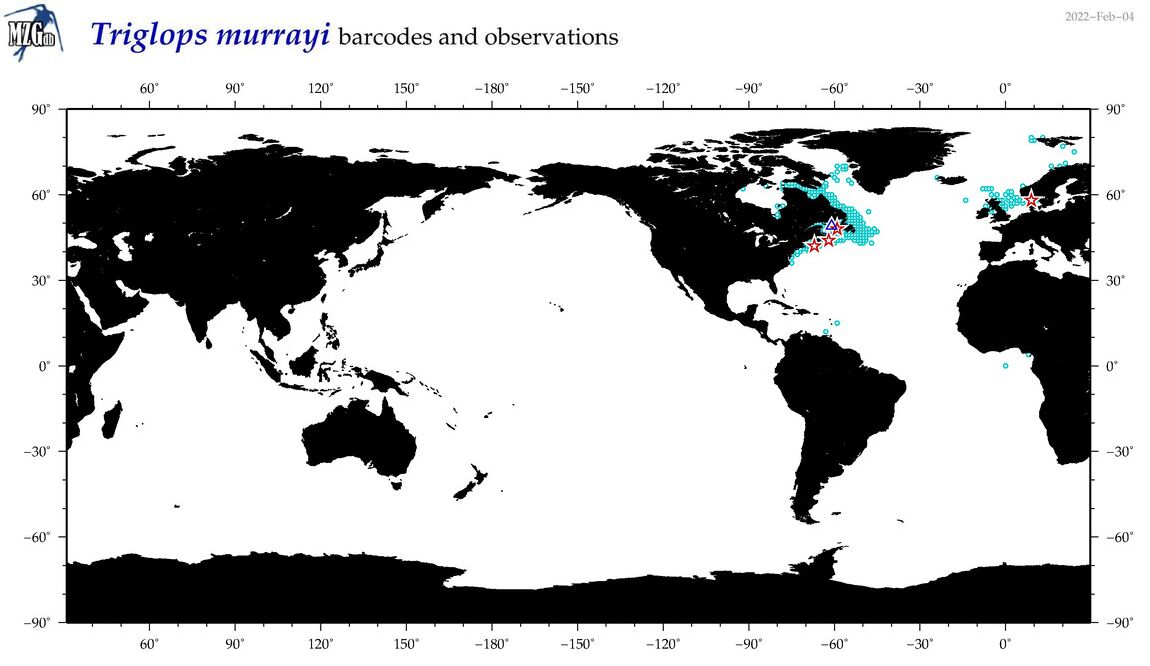 |
accepted
T5001373
R:1:0:0:0 |
| Trigonolampa miriceps |
Species
(44) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 2
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
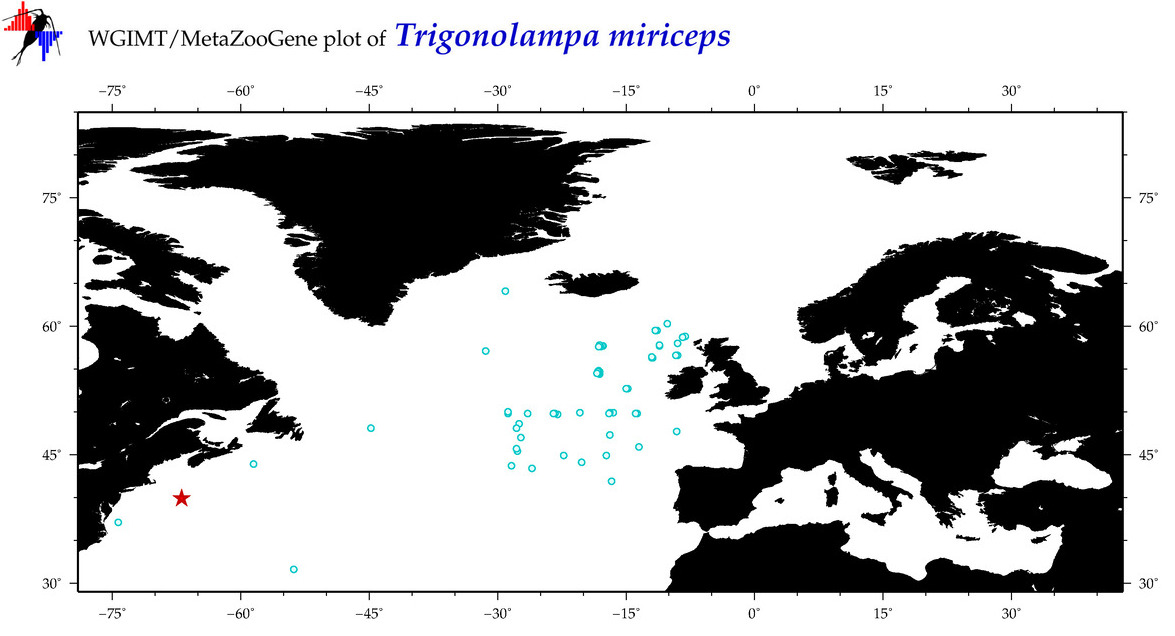 |
accepted
T5005798
R:1:0:0:0 |
| Zu cristatus |
Species
(45) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 5
16S = 3
18S = 0
28S = 1
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
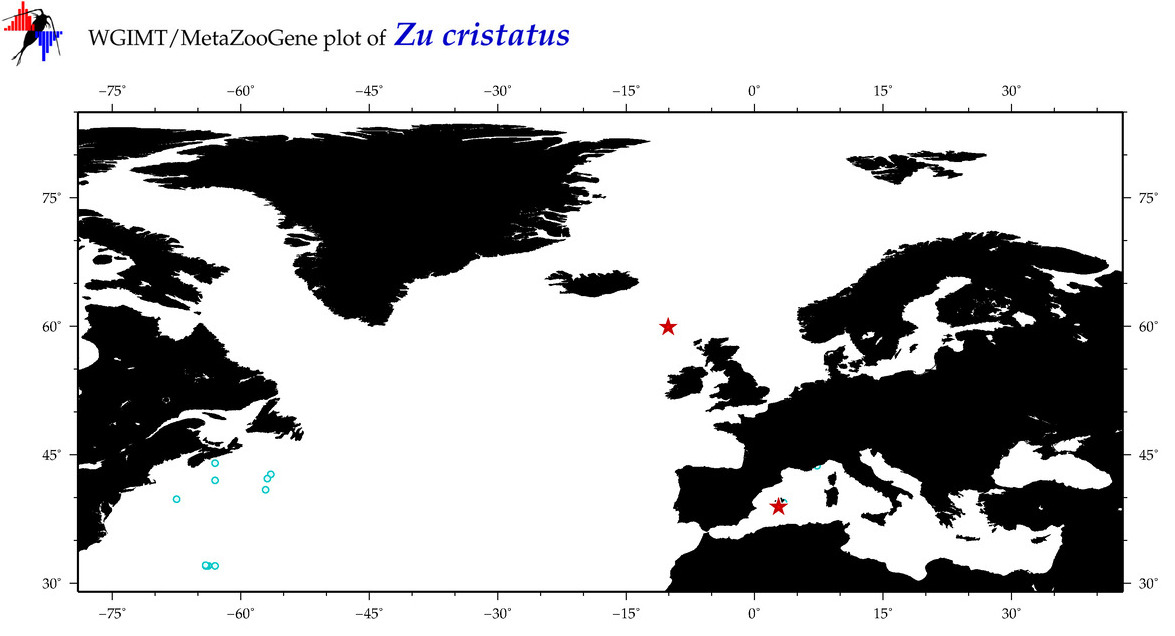 |
accepted
T5001425
R:1:0:0:0 |