Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-26 |
| Syngnathiformes |
Order |
cummulative
sub-totals
------->
for this
Order |
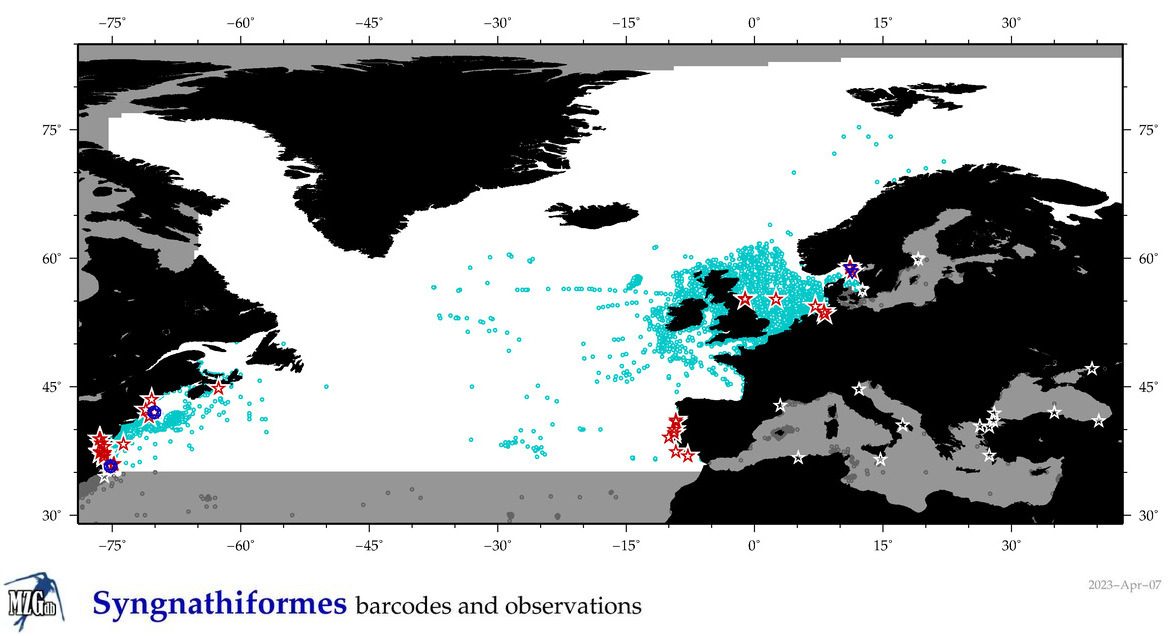
Total Species: 57
Sp. w/COI-Barcodes: 48
Total COI-Barcodes: 1686 |
Total Species: 57
Sp. w/COI-Barcodes: 31
Total COI-Barcodes: 193 |
 |
accepted
T5002224
R:1:1:1:0 |
| ⋄ Aulostomidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 45 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 11 |
 |
accepted
T5000237
R:1:0:0:0 |
| ⋄ ⋄ Aulostomus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 45 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 11 |
 |
accepted
T5002165
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Aulostomus chinensis |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 31
12S = 7
16S = 4
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003218
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Aulostomus maculatus |
Species
(2) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 2
16S = 4
18S = 1
28S = 1
|
COI = 11
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5002166
R:1:0:0:0 |
| ⋄ Centriscidae |
Family |
cummulative
sub-totals
------->
for this
Family |
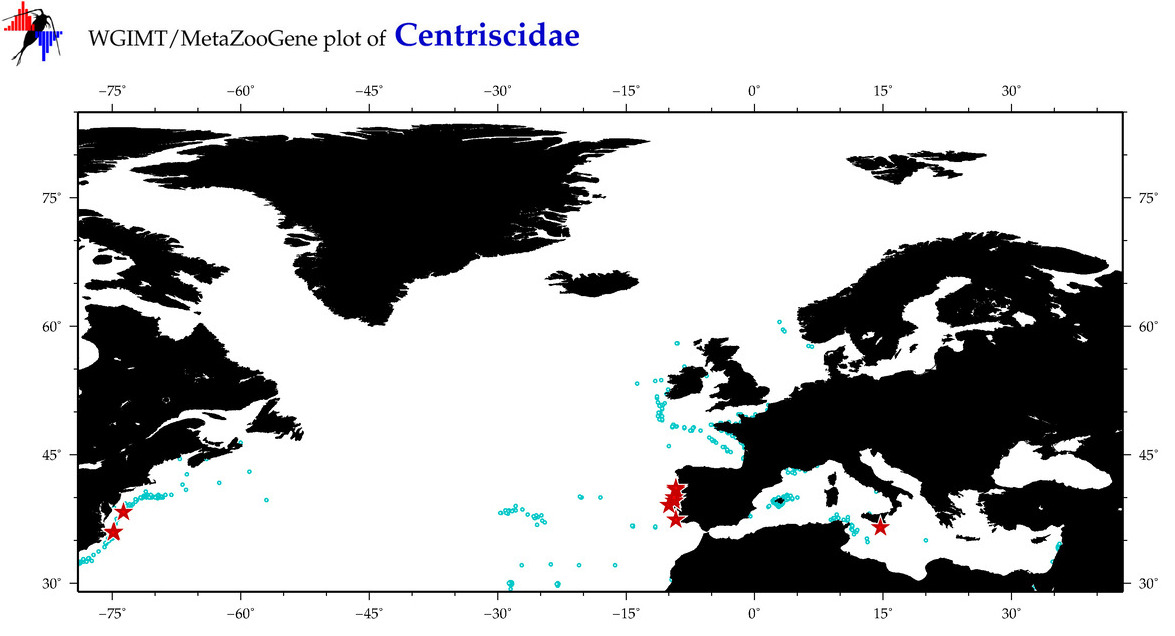
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 106 |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 16 |
 |
accepted
T5000355
R:1:1:0:0 |
| ⋄ ⋄ Centriscinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5006101
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Centriscus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 0
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5006338
R:1:1:0:0 |
| ⋄ ⋄ Macroramphosinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
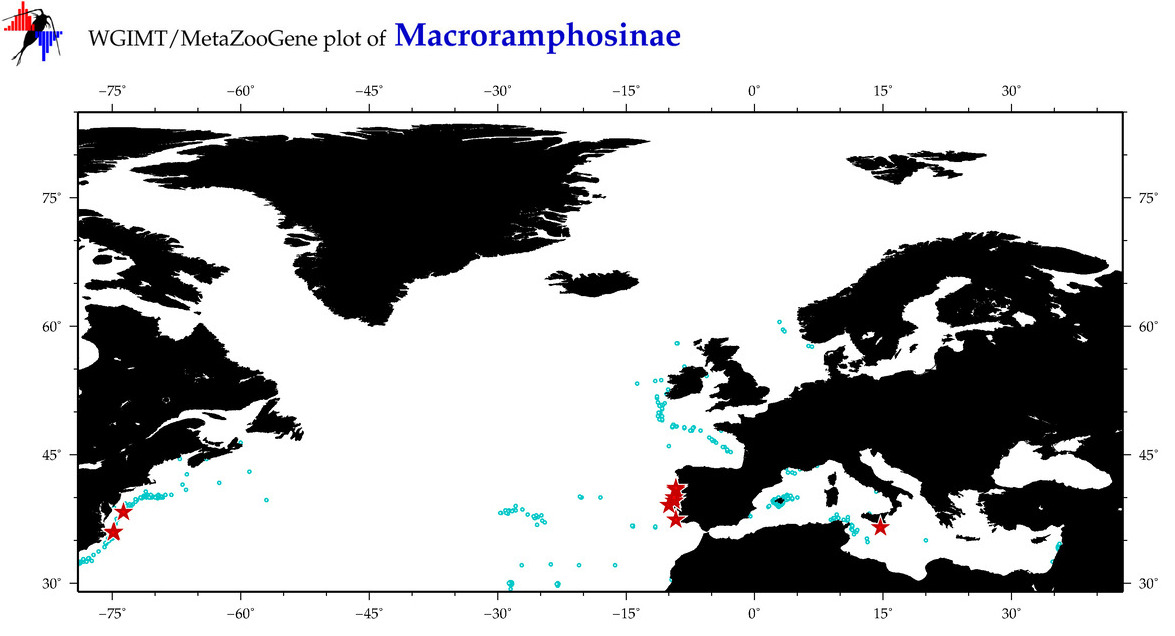
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 106 |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 16 |
 |
accepted
T5002355
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Macroramphosus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 104 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 16 |
 |
accepted
T5000857
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Macroramphosus gracilis |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 17
12S = 5
16S = 4
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
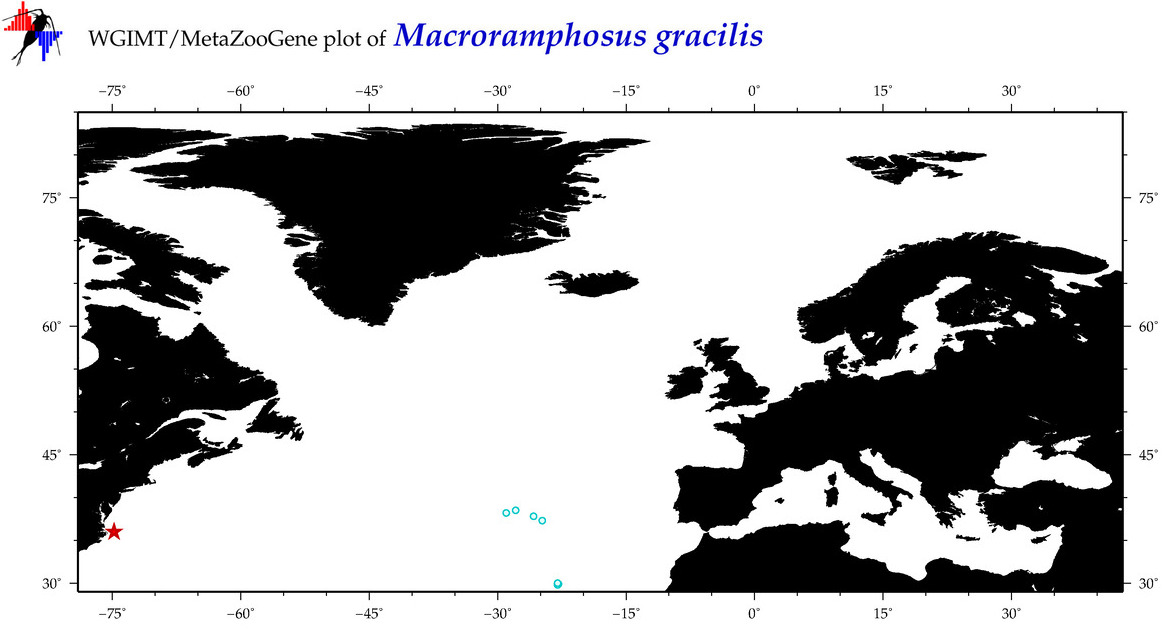 |
accepted
T5000856
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Macroramphosus scolopax |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 87
12S = 6
16S = 20
18S = 0
28S = 1
|
COI = 15
12S = 0
16S = 0
18S = 0
28S = 0
|
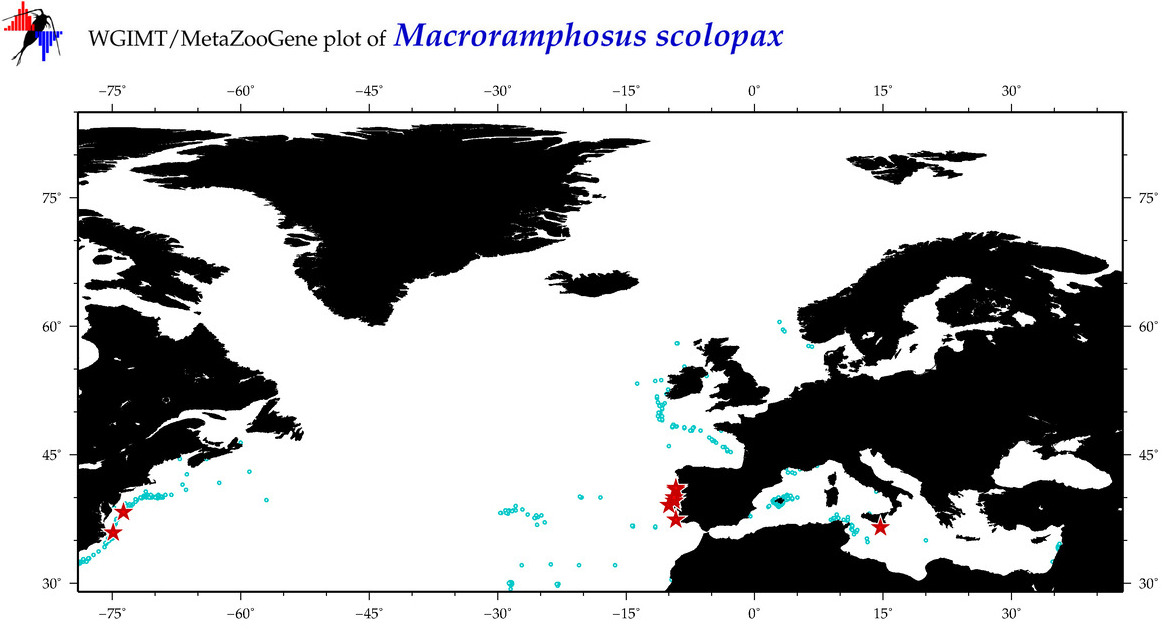 |
accepted
T5000858
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Notopogon |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
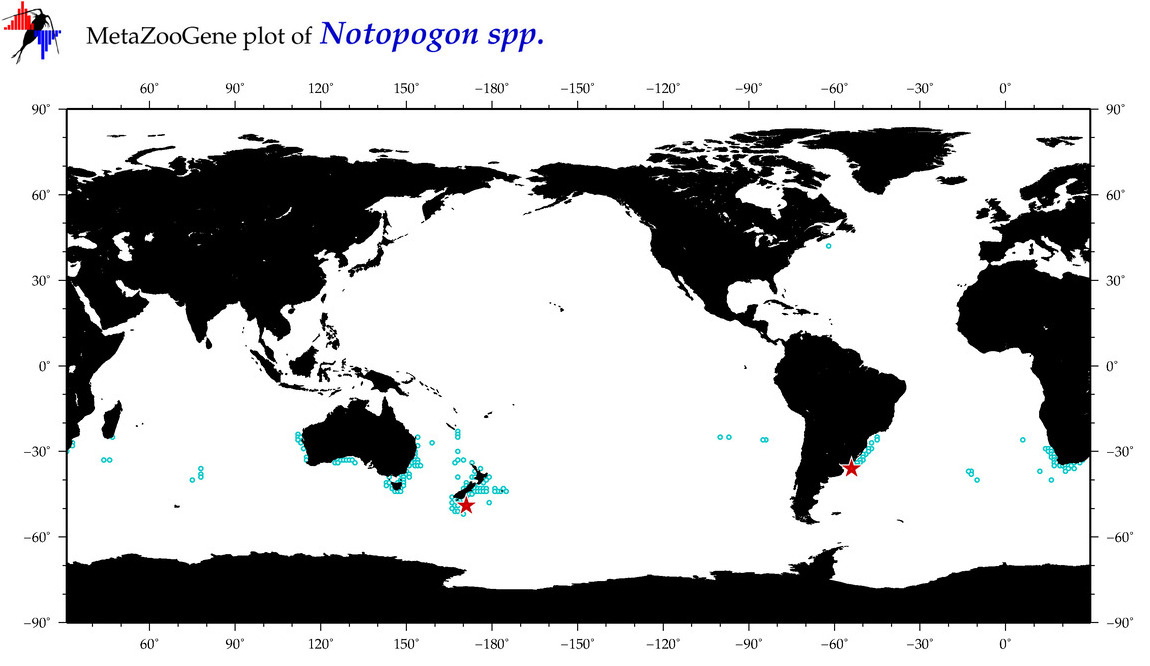 |
accepted
T5002798
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Notopogon fernandezianus |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
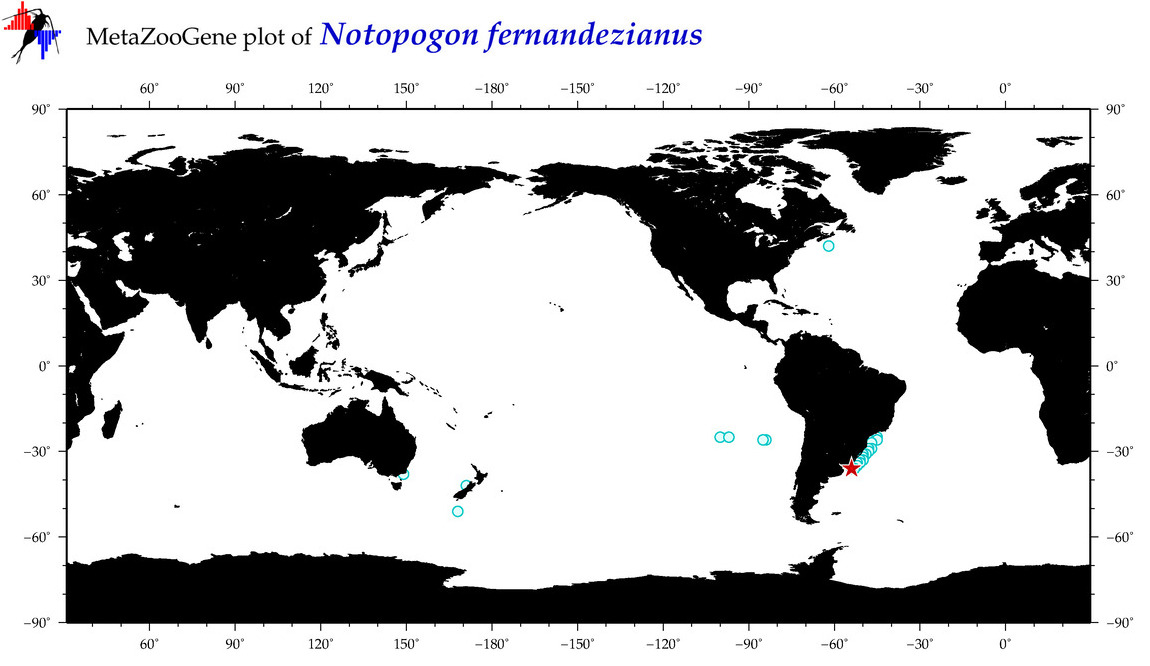 |
accepted
T5004814
R:1:0:0:0 |
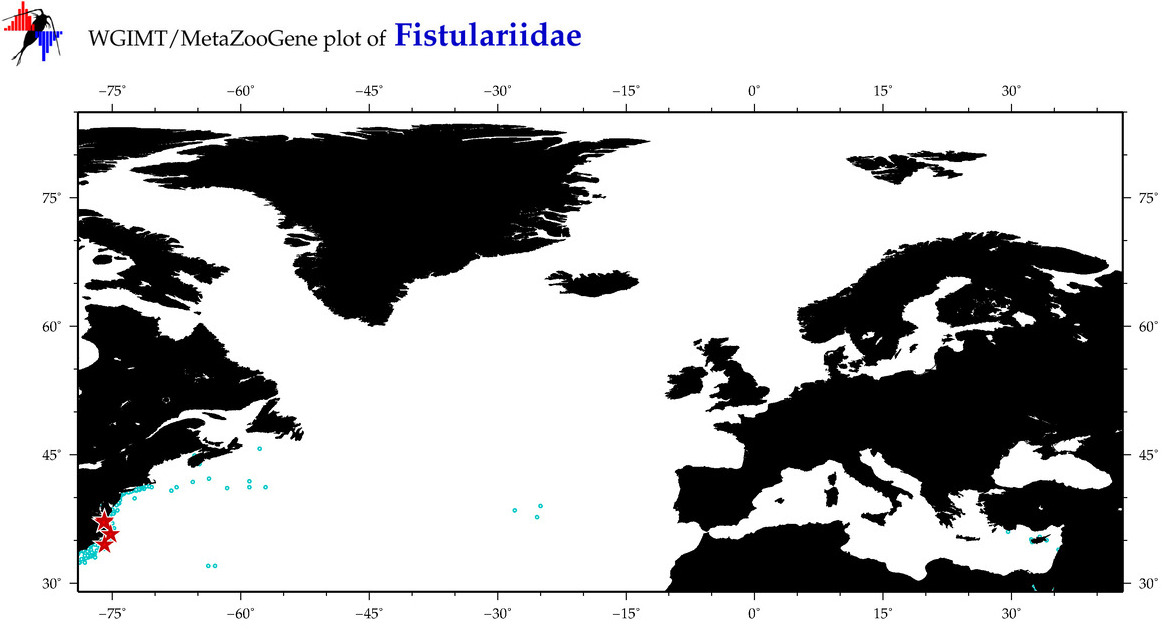
| ⋄ Fistulariidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 210 |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 7 |
 |
accepted
T5000584
R:1:1:0:0 |
| ⋄ ⋄ Fistularia |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 210 |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 7 |
 |
accepted
T5000583
R:1:1:0:0 |
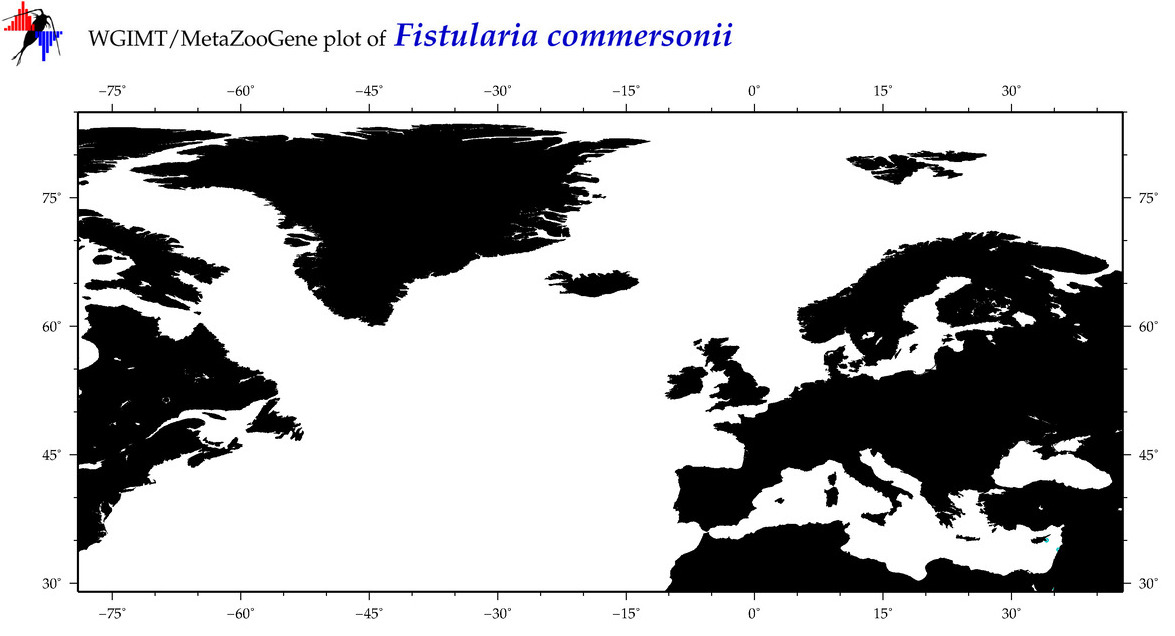
| ⋄ ⋄ ⋄ Fistularia commersonii |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 155
12S = 18
16S = 14
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003997
R:1:0:0:0 |
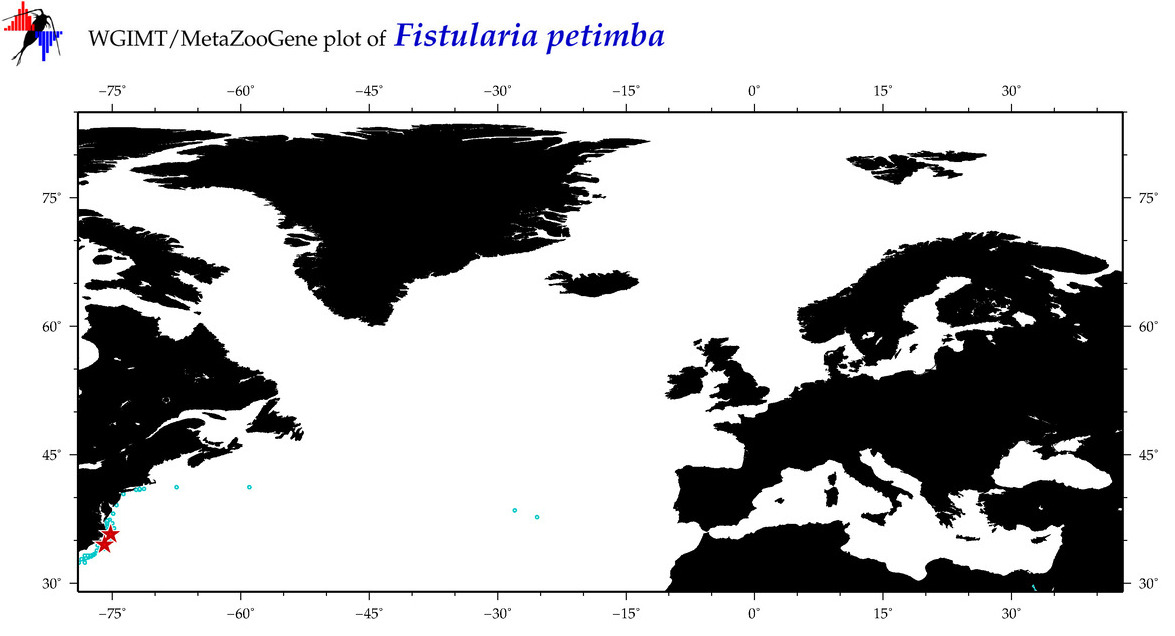
| ⋄ ⋄ ⋄ Fistularia petimba |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 45
12S = 15
16S = 11
18S = 1
28S = 0
|
COI = 3
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5003998
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Fistularia tabacaria |
Species
(8) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003999
R:1:1:0:0 |
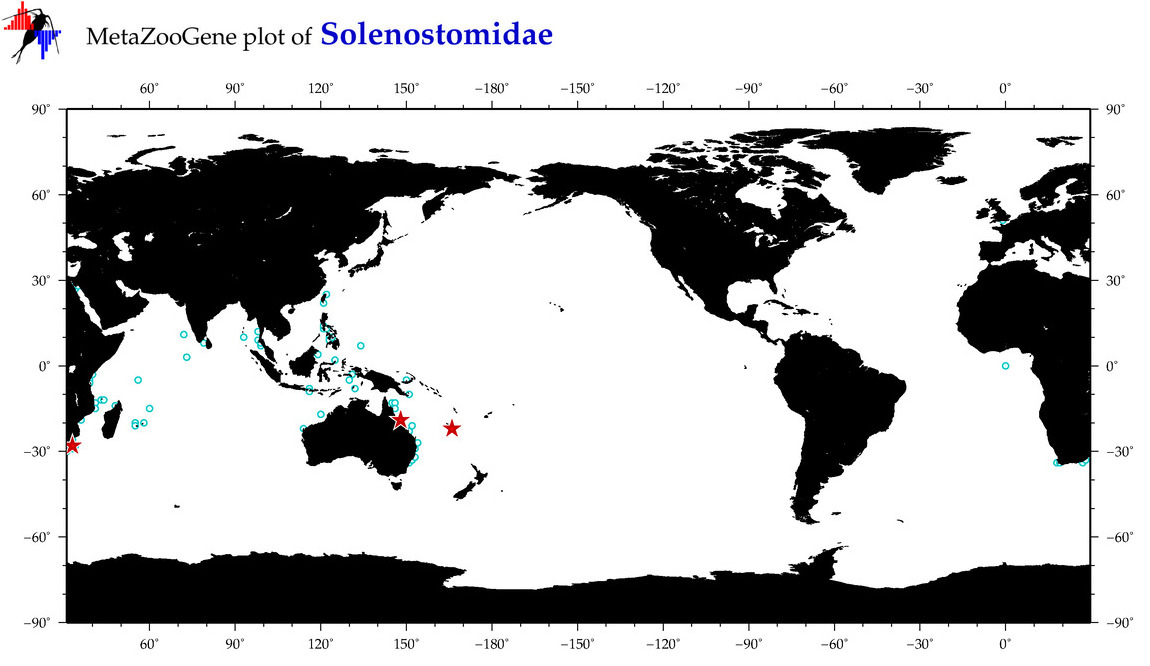
| ⋄ Solenostomidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 9 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5005992
R:1:0:0:0 |
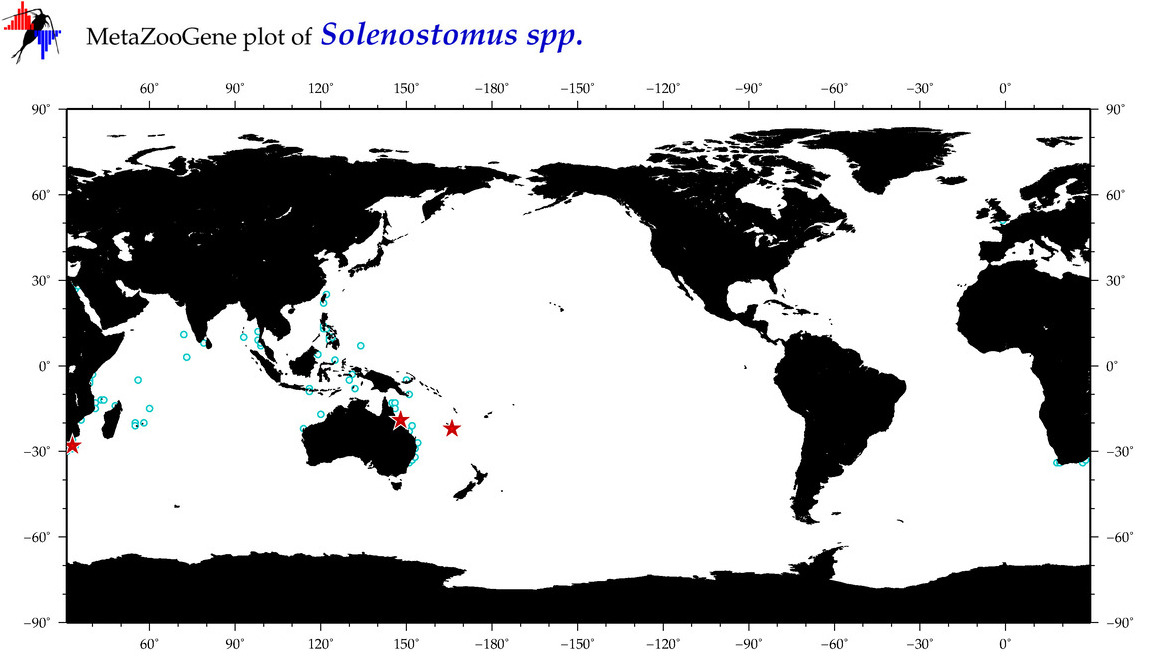
| ⋄ ⋄ Solenostomus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 9 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5006359
R:1:0:0:0 |
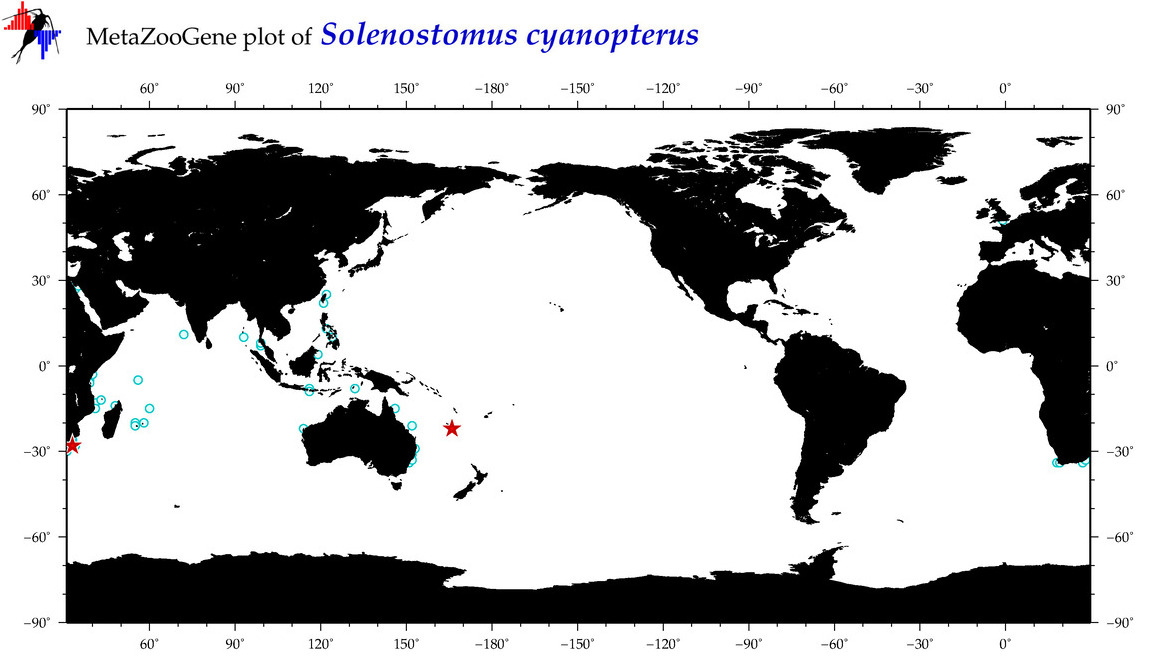
| ⋄ ⋄ ⋄ Solenostomus cyanopterus |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 9
12S = 5
16S = 3
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5012272
R:1:0:0:0 |
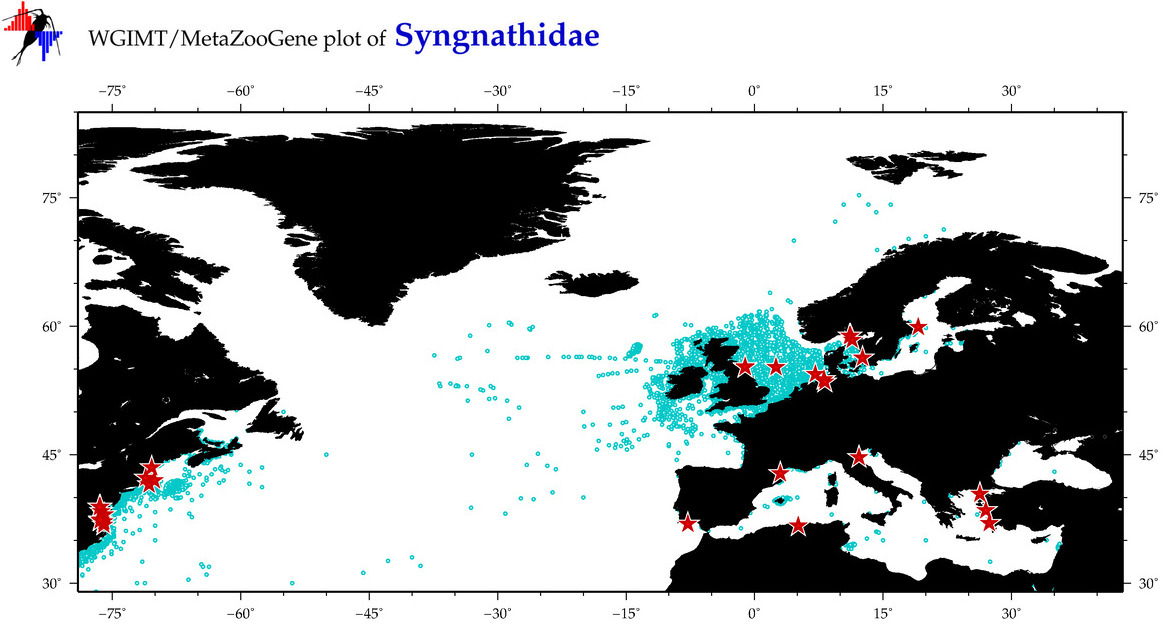
| ⋄ Syngnathidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 48
Sp. w/COI-Barcodes: 39
Total COI-Barcodes: 1316 |
Total Species: 48
Sp. w/COI-Barcodes: 26
Total COI-Barcodes: 159 |
 |
accepted
T5000121
R:1:1:1:0 |
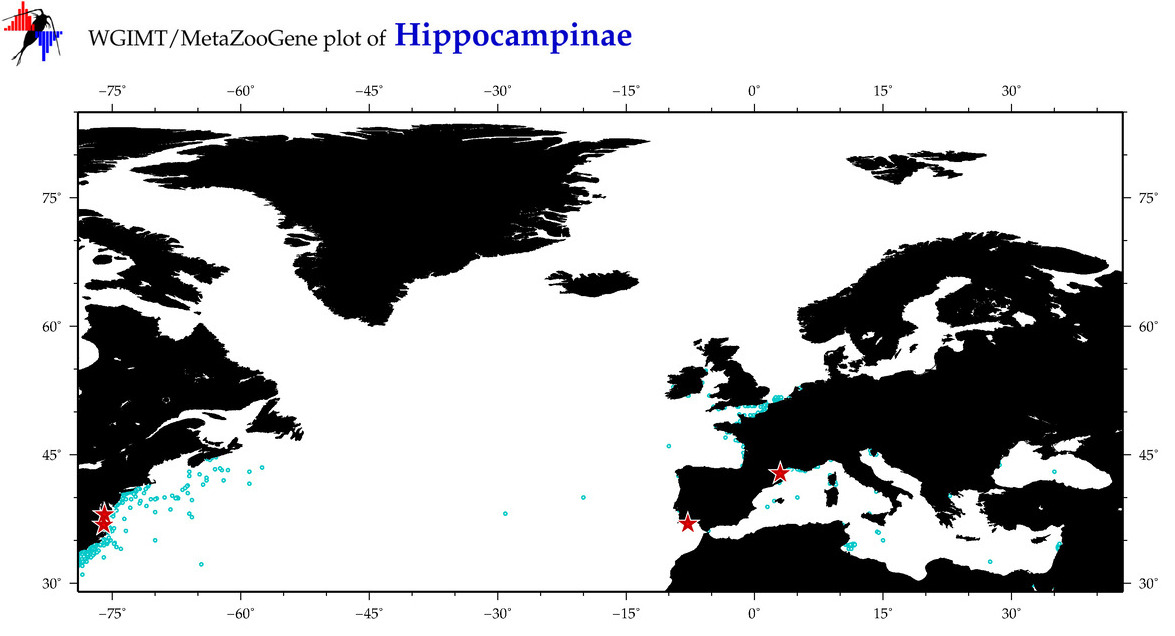
| ⋄ ⋄ Hippocampinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 11
Sp. w/COI-Barcodes: 11
Total COI-Barcodes: 696 |
Total Species: 11
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 19 |
 |
accepted
T5002357
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Hippocampus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 11
Sp. w/COI-Barcodes: 11
Total COI-Barcodes: 696 |
Total Species: 11
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 19 |
 |
accepted
T5000668
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus algiricus |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 2
16S = 4
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004251
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus breviceps |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 24
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5020853
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus comes |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 87
12S = 2
16S = 8
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
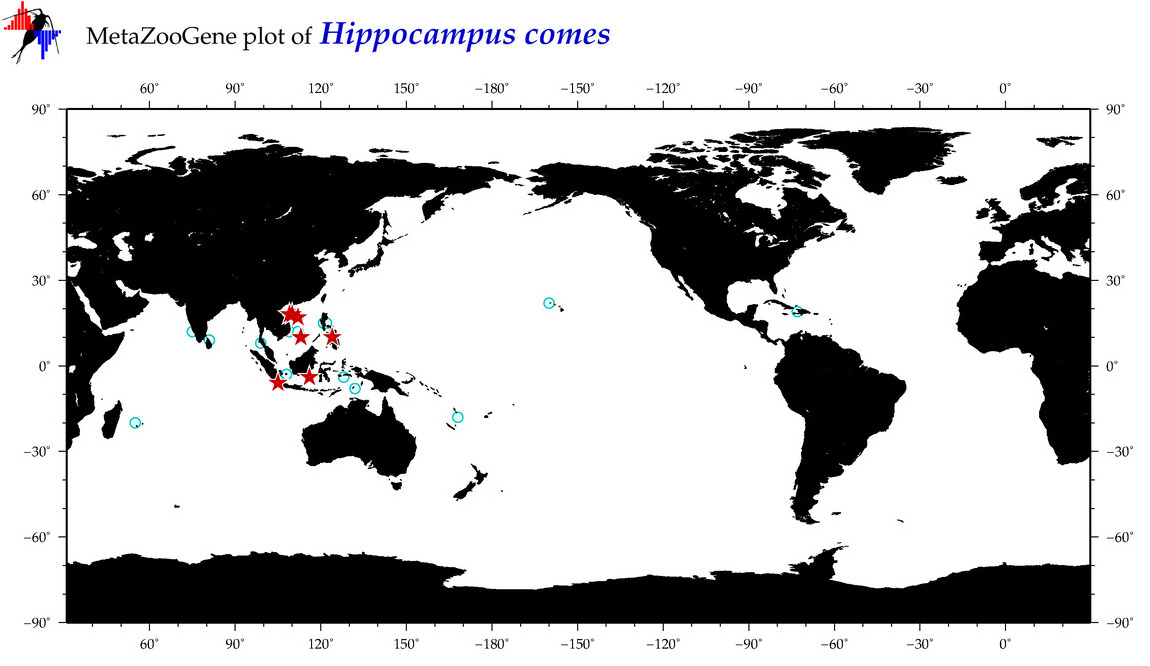 |
accepted
T5004258
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus erectus |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 49
12S = 5
16S = 18
18S = 0
28S = 0
|
COI = 13
12S = 0
16S = 0
18S = 0
28S = 0
|
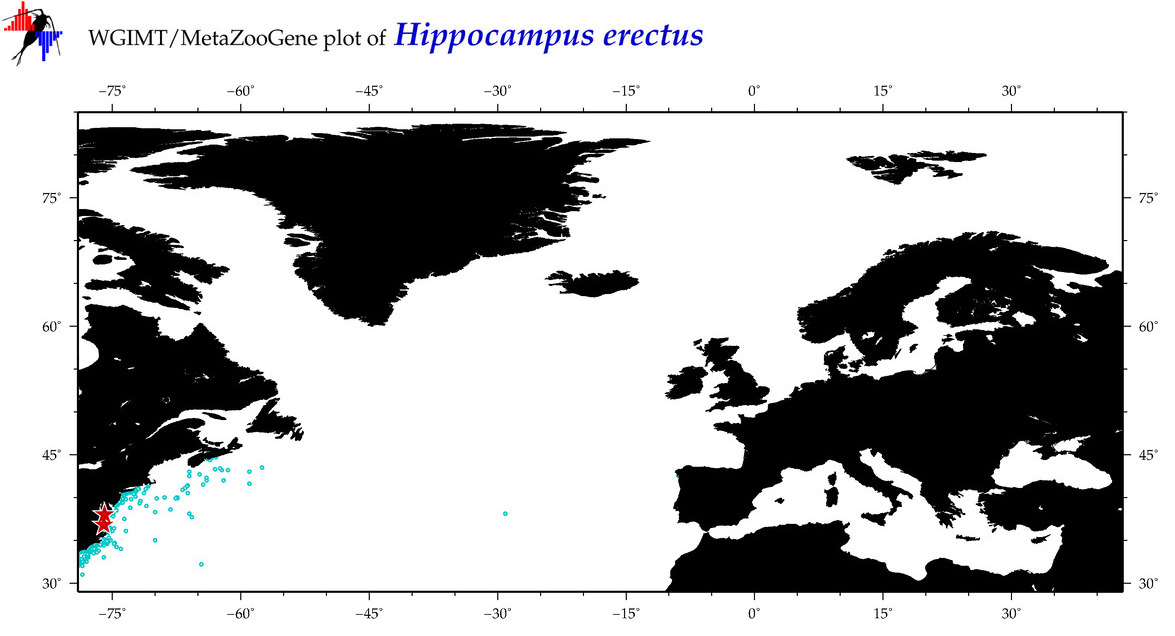 |
accepted
T5000669
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus guttulatus |
Species
(14) |
COI
12S
16S
18S
28S
|
COI = 12
12S = 1
16S = 8
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
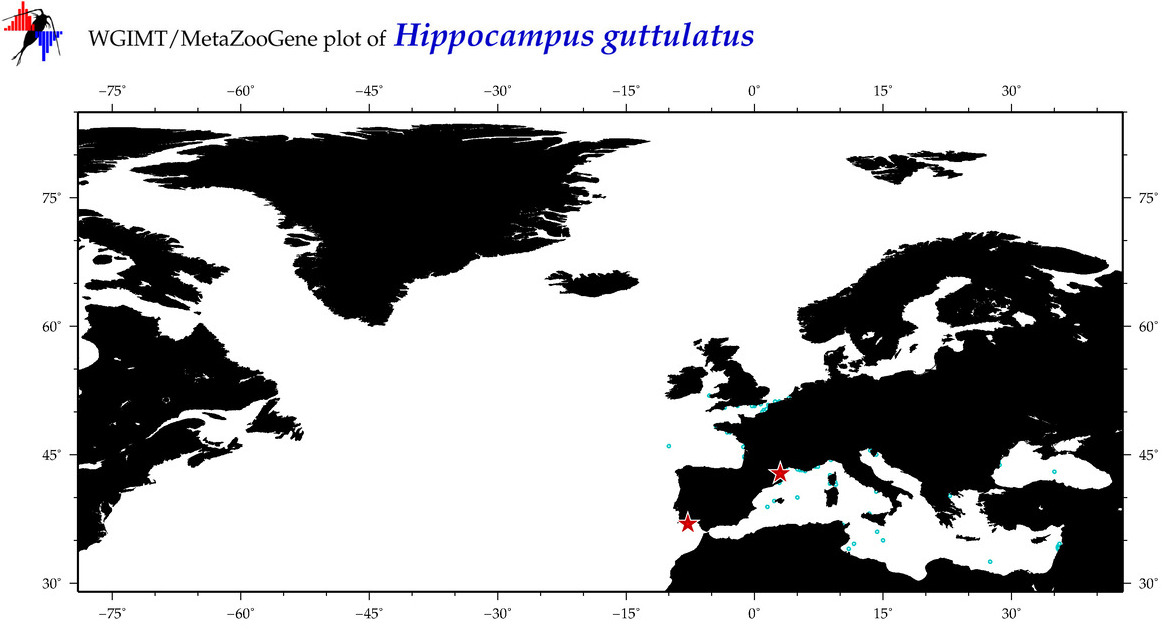 |
accepted
T5004262
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus hippocampus |
Species
(15) |
COI
12S
16S
18S
28S
|
COI = 48
12S = 3
16S = 24
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
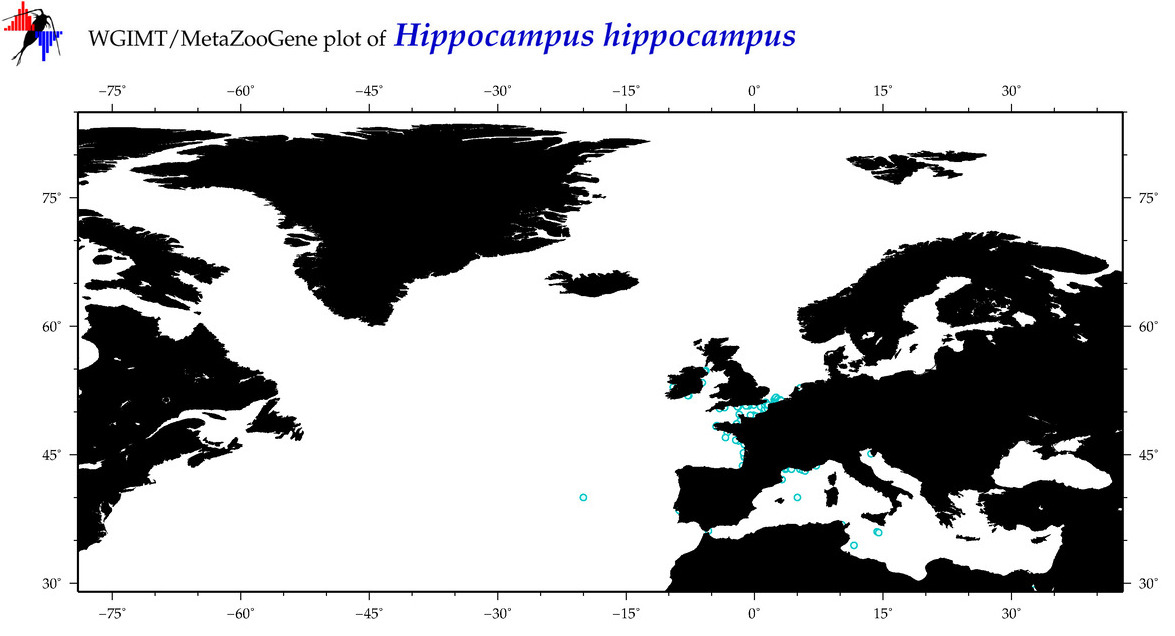 |
accepted
T5004264
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus ingens |
Species
(16) |
COI
12S
16S
18S
28S
|
COI = 65
12S = 3
16S = 7
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004266
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus kelloggi |
Species
(17) |
COI
12S
16S
18S
28S
|
COI = 66
12S = 3
16S = 6
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
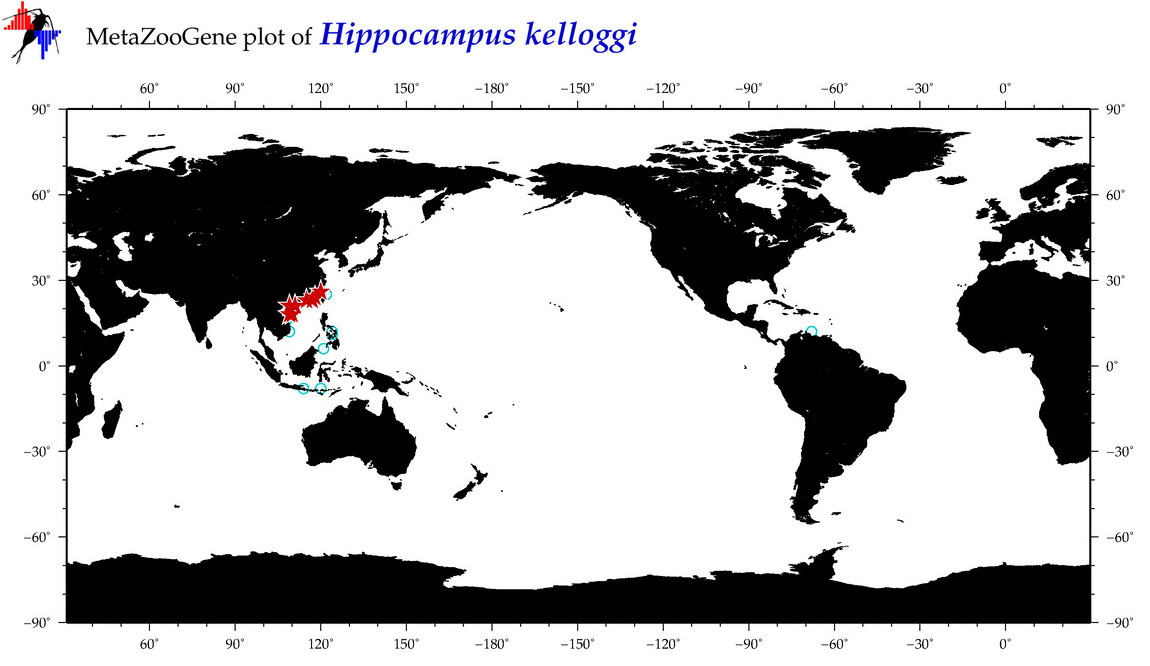 |
accepted
T5004268
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus kuda |
Species
(18) |
COI
12S
16S
18S
28S
|
COI = 140
12S = 10
16S = 45
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004269
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus reidi |
Species
(19) |
COI
12S
16S
18S
28S
|
COI = 54
12S = 4
16S = 9
18S = 1
28S = 0
|
COI = 3
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5004274
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus zosterae |
Species
(20) |
COI
12S
16S
18S
28S
|
COI = 147
12S = 5
16S = 5
18S = 1
28S = 0
|
COI = 2
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5004279
R:1:0:0:0 |
| ⋄ ⋄ Syngnathinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 37
Sp. w/COI-Barcodes: 28
Total COI-Barcodes: 620 |
Total Species: 37
Sp. w/COI-Barcodes: 22
Total COI-Barcodes: 140 |
 |
accepted
T5002356
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Acentronura |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
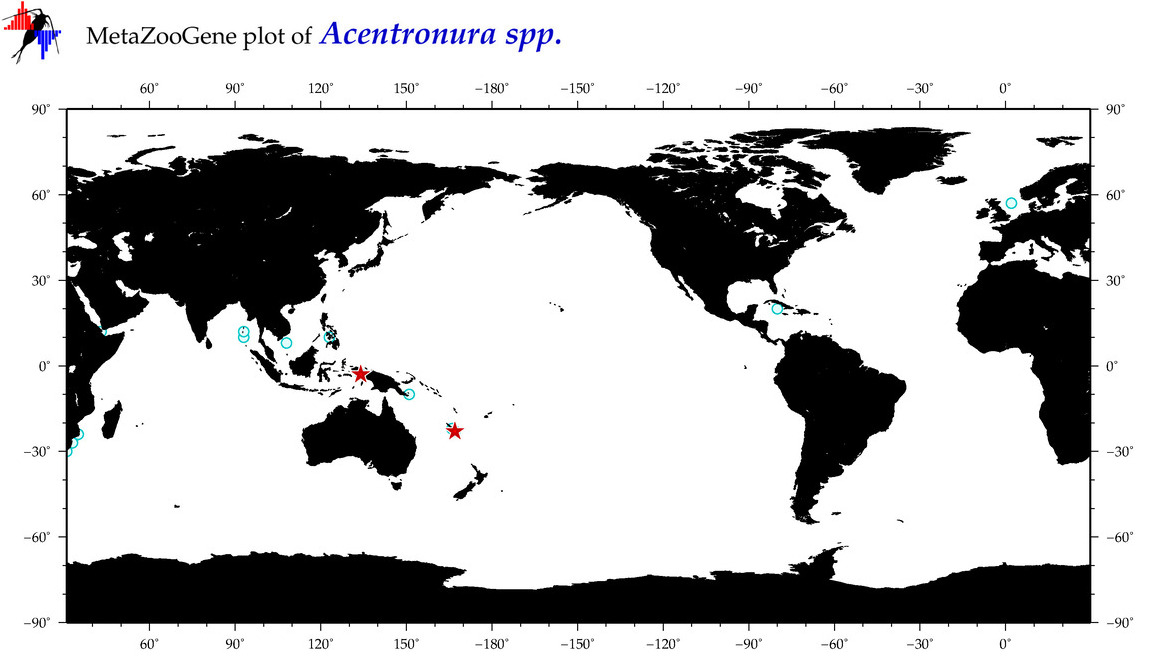 |
accepted
T5002417
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Acentronura tentaculata |
Species
(21) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 3
16S = 3
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
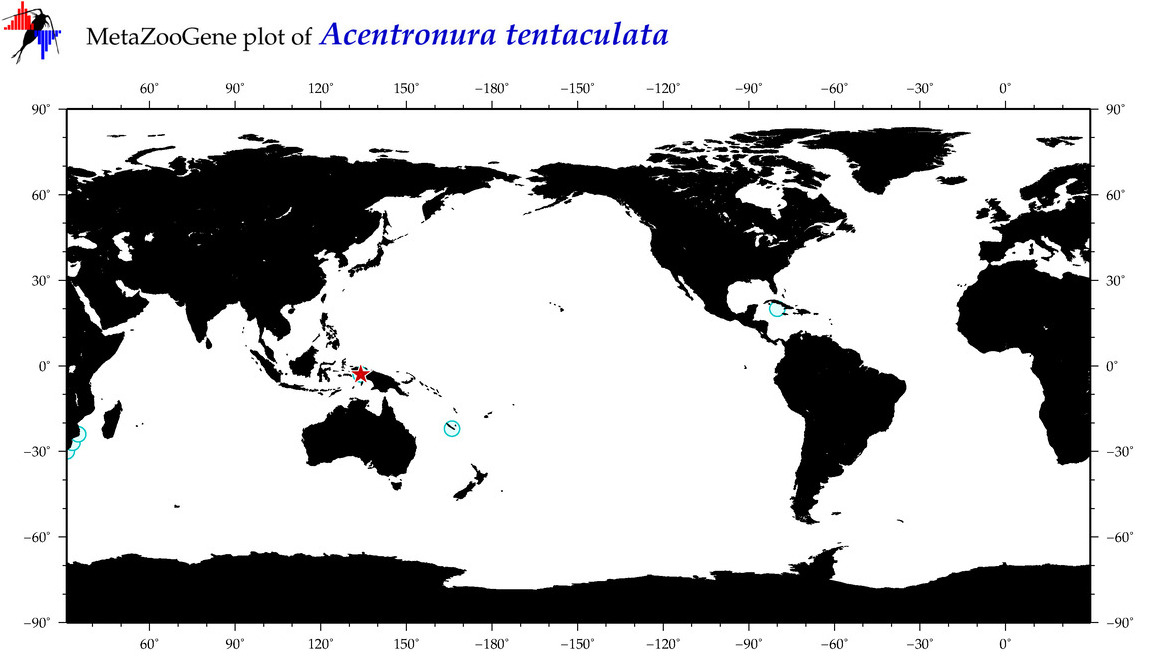 |
accepted
T5003032
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Amphelikturus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 3 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 3 |
 |
accepted
T5002440
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Amphelikturus dendriticus |
Species
(22) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 3
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5003086
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Anarchopterus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 7 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 7 |
 |
accepted
T5002441
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Anarchopterus criniger |
Species
(23) |
COI
12S
16S
18S
28S
|
COI = 7
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 7
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5003088
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Anarchopterus tectus |
Species
(24) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5016254
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Bryx |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 25 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 24 |
 |
accepted
T5002498
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Bryx dunckeri |
Species
(25) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 2
16S = 2
18S = 1
28S = 0
|
COI = 19
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5003319
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Bryx randalli |
Species
(26) |
COI
12S
16S
18S
28S
|
COI = 5
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 5
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5003320
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Corythoichthys |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 69 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002545
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Corythoichthys flavofasciatus |
Species
(27) |
COI
12S
16S
18S
28S
|
COI = 69
12S = 2
16S = 3
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
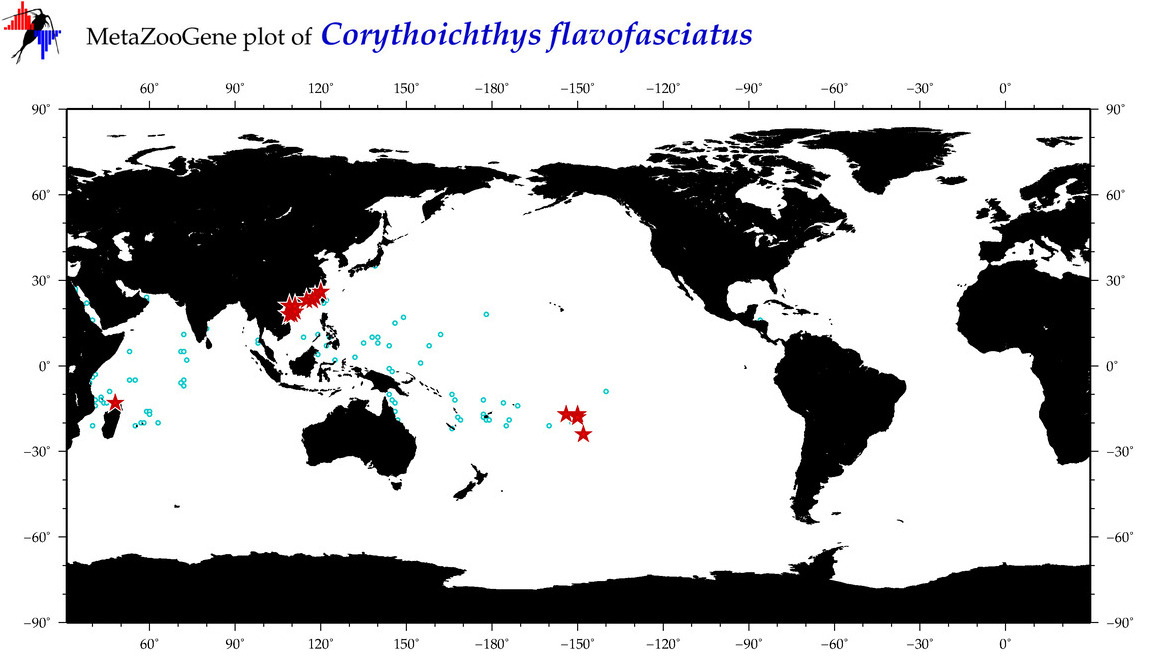 |
accepted
T5003645
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Cosmocampus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 6
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 18 |
Total Species: 6
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 11 |
 |
accepted
T5002546
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cosmocampus albirostris |
Species
(28) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 4
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5003649
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cosmocampus brachycephalus |
Species
(29) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
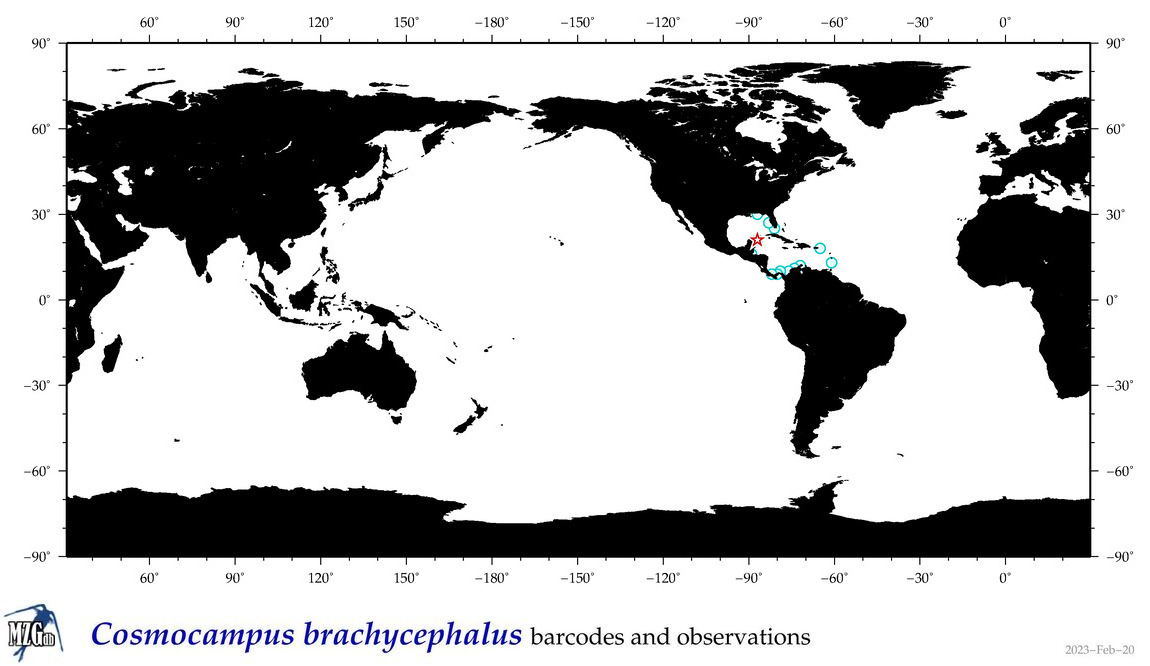 |
accepted
T5018679
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cosmocampus elucens |
Species
(30) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 3
16S = 3
18S = 1
28S = 0
|
COI = 6
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5003653
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cosmocampus hildebrandi |
Species
(31) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5018681
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cosmocampus profundus |
Species
(32) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5018683
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Cosmocampus retropinnis |
Species
(33) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5018684
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Enneacampus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
Total Species: 2
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002602
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Entelurus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 10 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
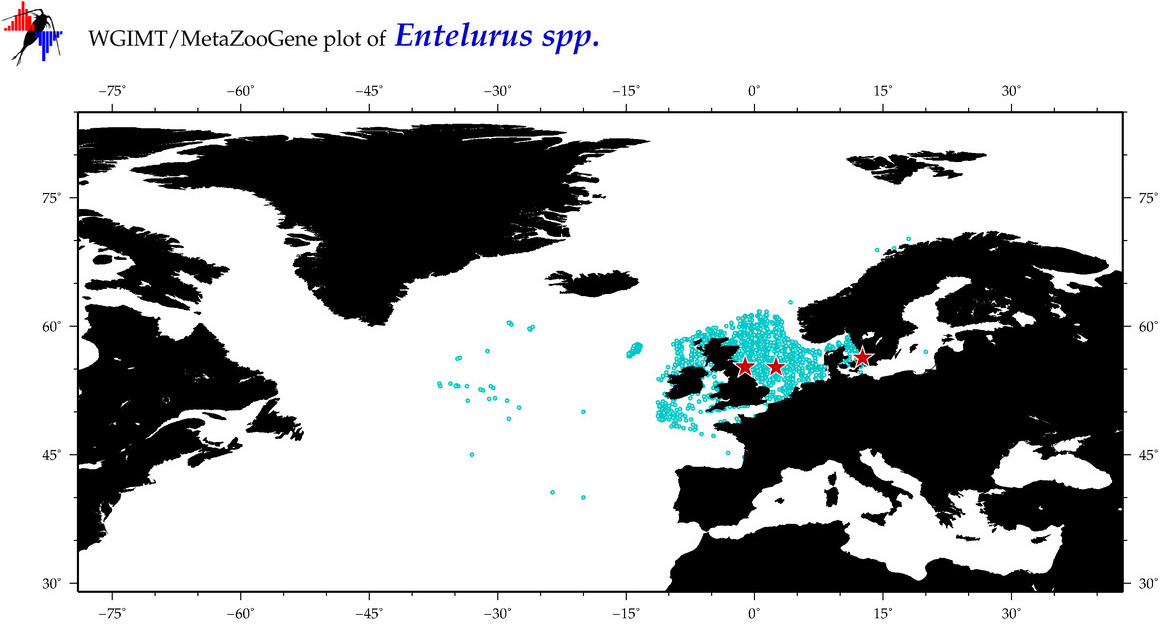 |
accepted
T5002603
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Entelurus aequoreus |
Species
(34) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 3
16S = 5
18S = 1
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
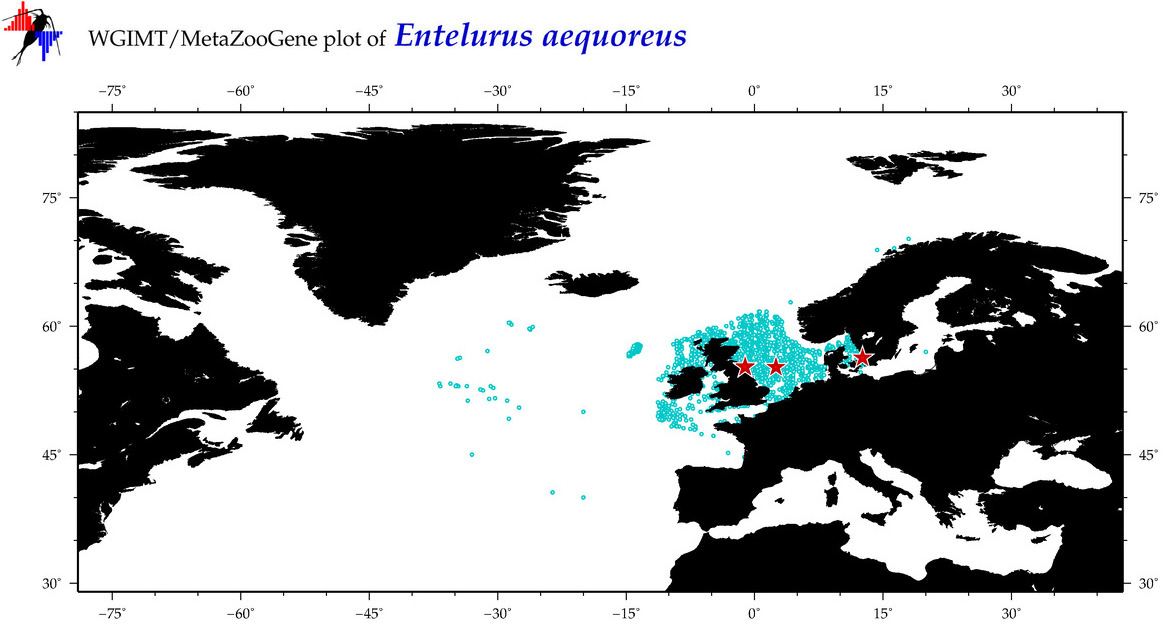 |
accepted
T5003877
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Micrognathus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 1 |
 |
accepted
T5002762
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Micrognathus crinitus |
Species
(35) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 1
12S = 1
16S = 1
18S = 1
28S = 0
|
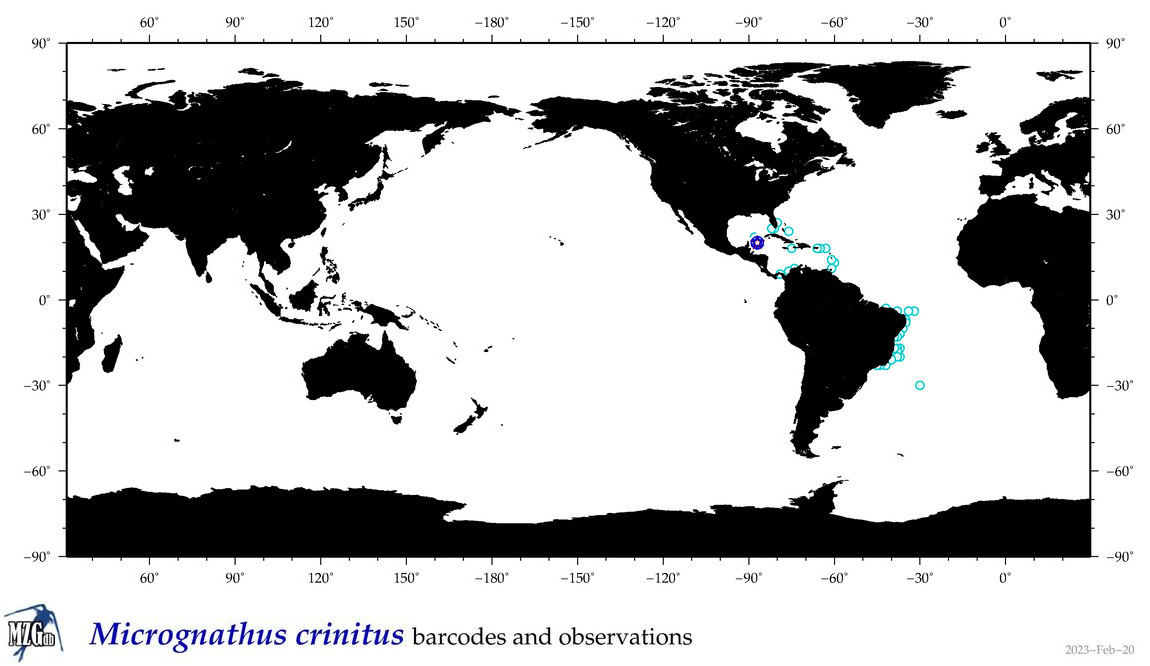 |
accepted
T5022182
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Microphis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 114 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 7 |
 |
accepted
T5002765
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Microphis brachyurus |
Species
(36) |
COI
12S
16S
18S
28S
|
COI = 106
12S = 8
16S = 4
18S = 1
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5022195
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Microphis lineatus |
Species
(37) |
COI
12S
16S
18S
28S
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004681
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Nerophis |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 3
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 5 |
Total Species: 3
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002792
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Nerophis lumbriciformis |
Species
(38) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 0
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004791
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Nerophis ophidion |
Species
(39) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 3
16S = 4
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004792
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Penetopteryx |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 2 |
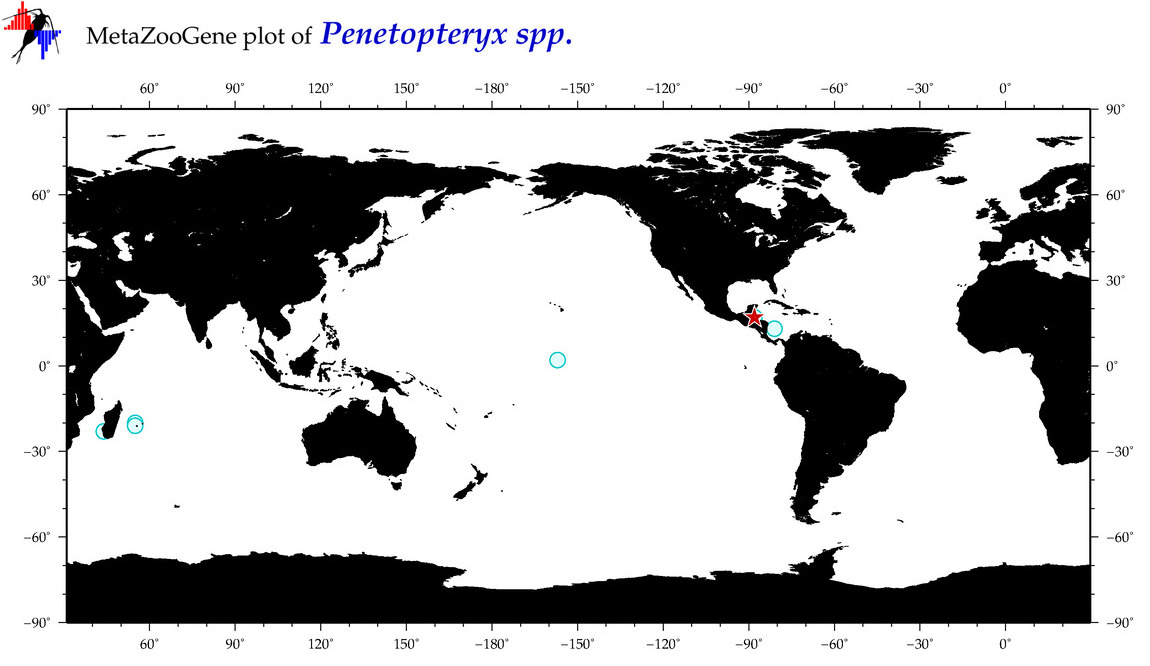 |
accepted
T5002841
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Penetopteryx nanus |
Species
(40) |
COI
12S
16S
18S
28S
|
COI = 2
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 2
12S = 1
16S = 1
18S = 1
28S = 0
|
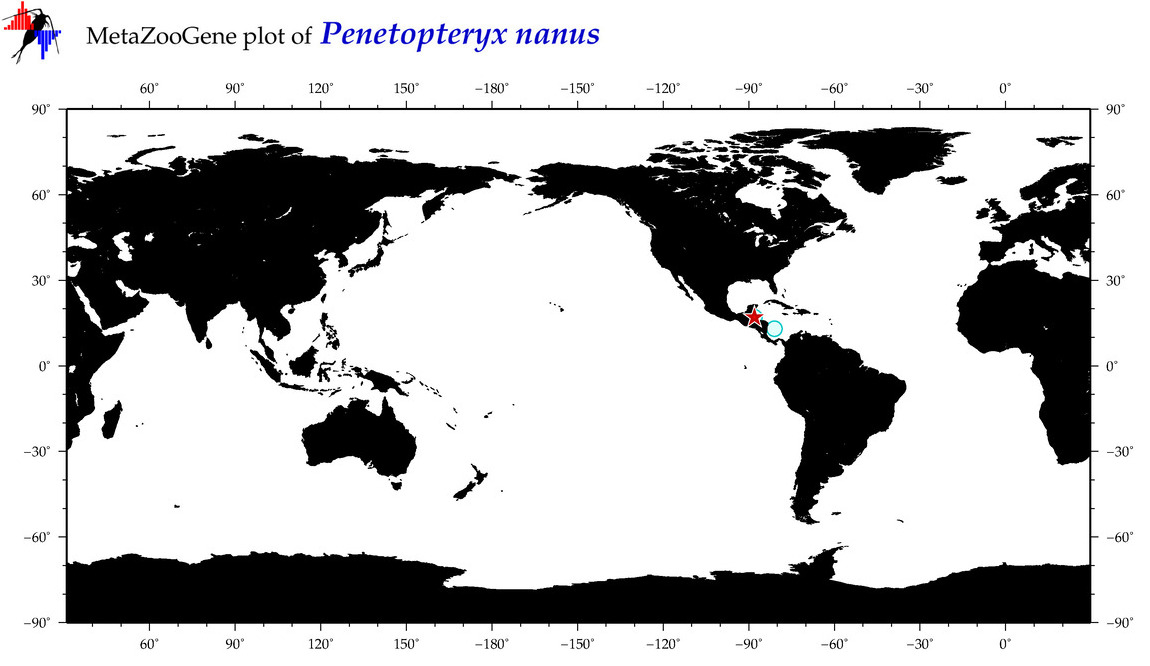 |
accepted
T5004999
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Phyllopteryx |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 144 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
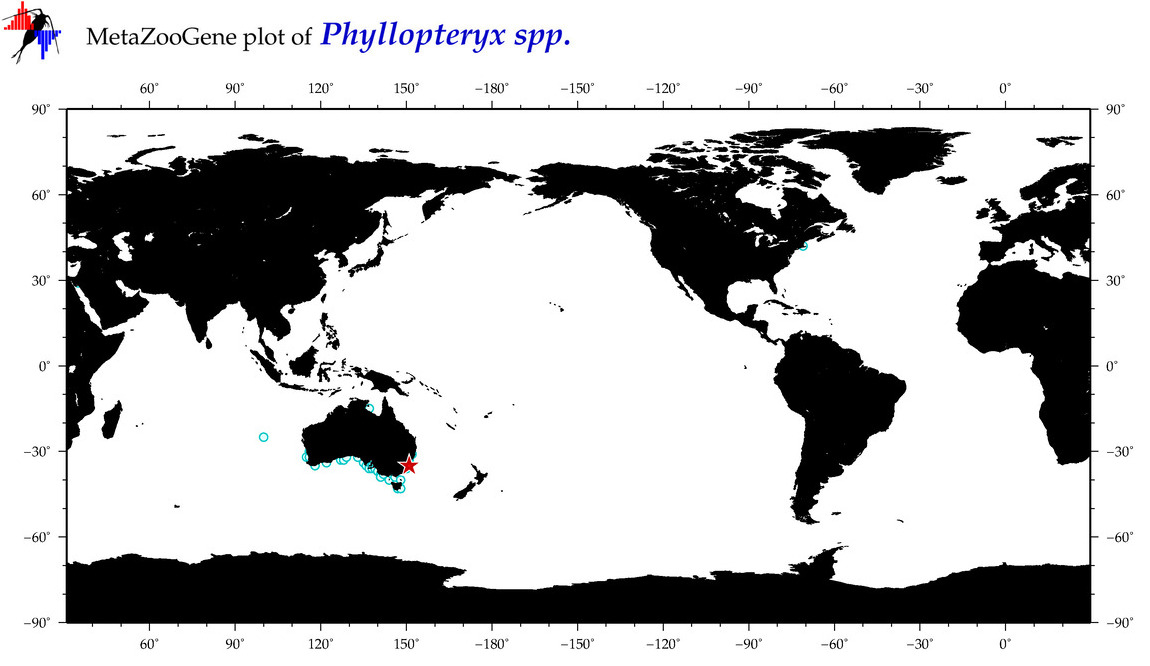 |
accepted
T5002855
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Phyllopteryx taeniolatus |
Species
(41) |
COI
12S
16S
18S
28S
|
COI = 144
12S = 8
16S = 8
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
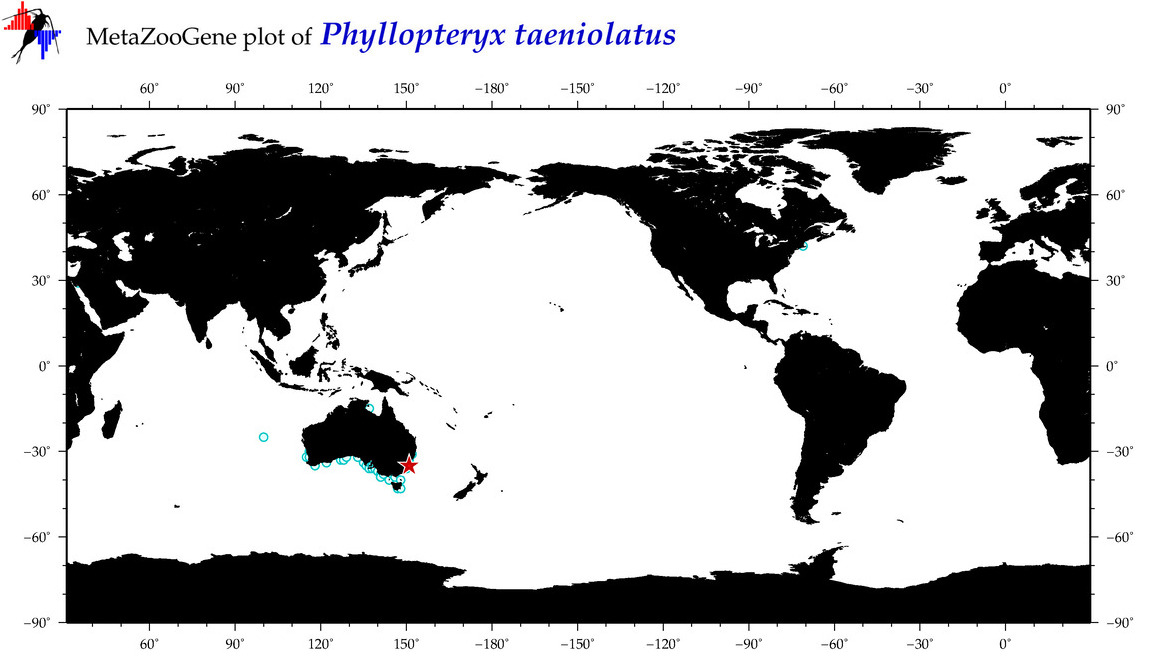 |
accepted
T5005040
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Pseudophallus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 6 |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 6 |
 |
accepted
T5002883
R:0:1:1:0 |
| ⋄ ⋄ ⋄ Syngnathus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 12
Sp. w/COI-Barcodes: 10
Total COI-Barcodes: 212 |
Total Species: 12
Sp. w/COI-Barcodes: 9
Total COI-Barcodes: 75 |
 |
accepted
T5001322
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus abaster |
Species
(42) |
COI
12S
16S
18S
28S
|
COI = 33
12S = 123
16S = 124
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
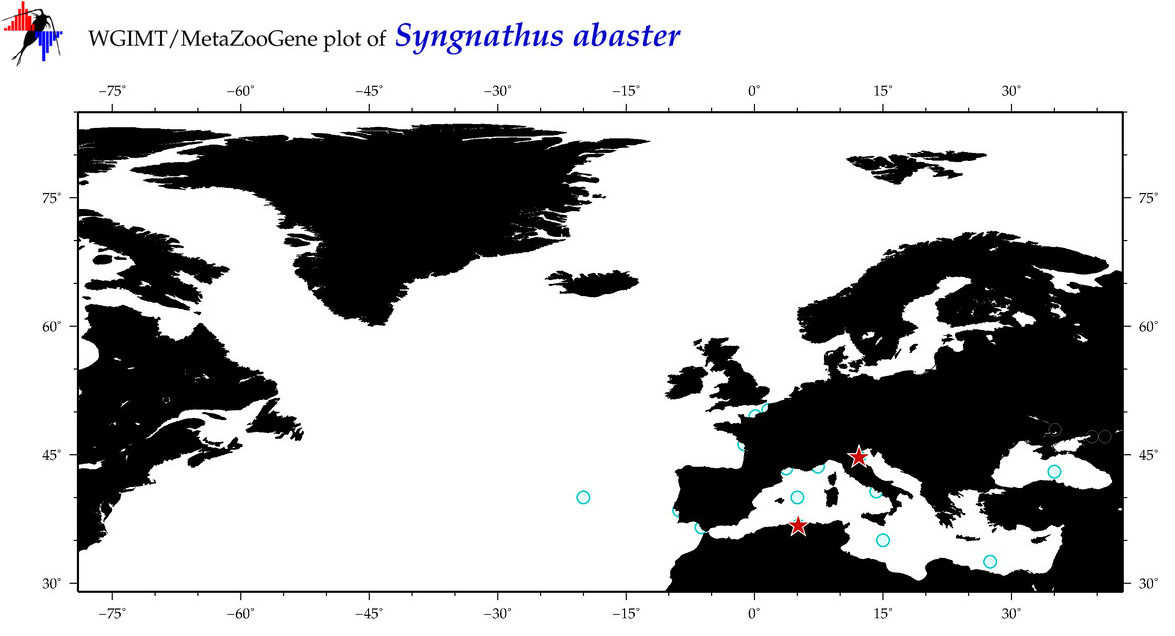 |
accepted
T5005672
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus acus |
Species
(43) |
COI
12S
16S
18S
28S
|
COI = 42
12S = 13
16S = 10
18S = 0
28S = 0
|
COI = 2
12S = 0
16S = 1
18S = 0
28S = 0
|
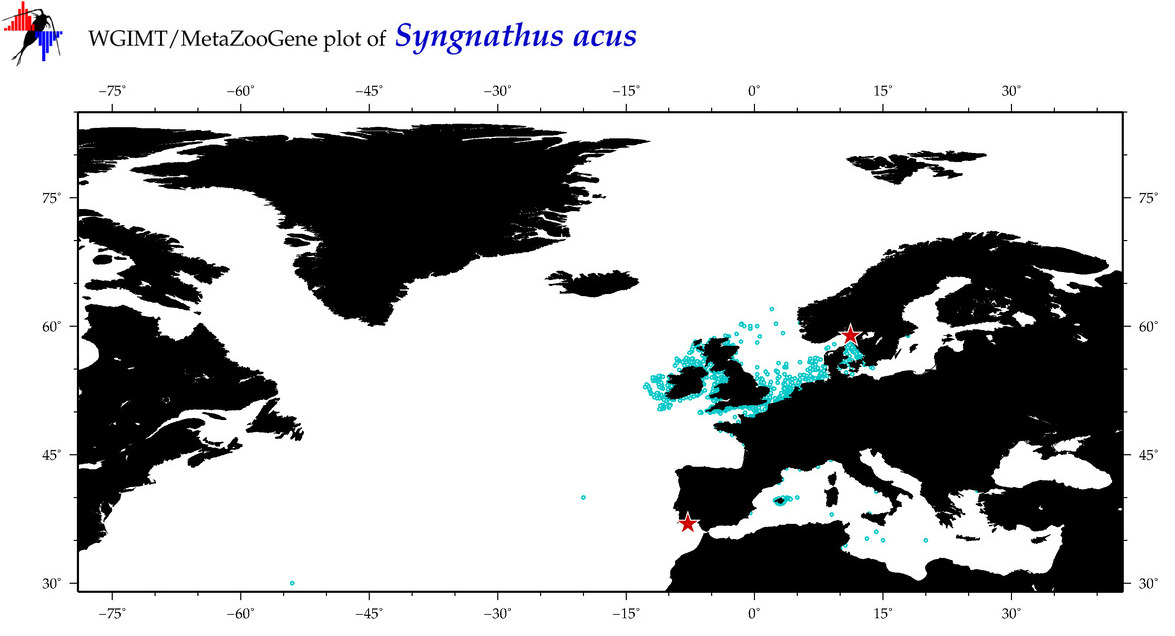 |
accepted
T5005673
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus affinis |
Species
(44) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026066
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus caribbaeus |
Species
(45) |
COI
12S
16S
18S
28S
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005675
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus dawsoni |
Species
(46) |
No barcodes? |
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026068
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus floridae |
Species
(47) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 5
16S = 5
18S = 0
28S = 0
|
COI = 17
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005678
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus fuscus |
Species
(48) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 3
16S = 2
18S = 1
28S = 0
|
COI = 17
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5005679
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus louisianae |
Species
(49) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 3
16S = 3
18S = 1
28S = 0
|
COI = 14
12S = 1
16S = 1
18S = 1
28S = 0
|
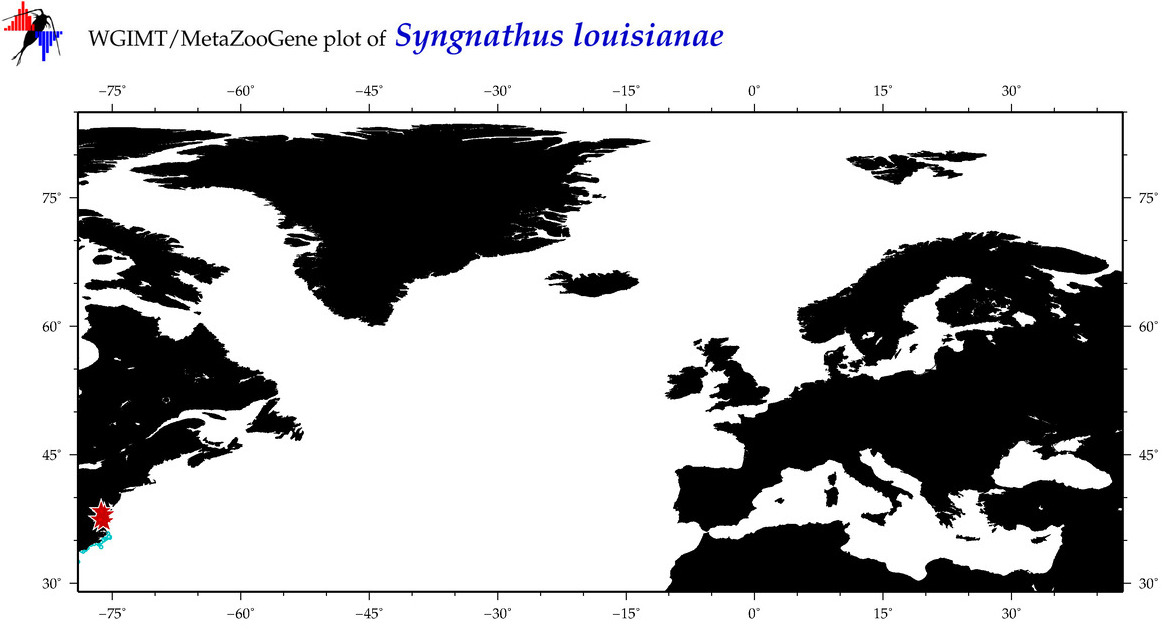 |
accepted
T5001325
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus rostellatus |
Species
(50) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 3
16S = 6
18S = 0
28S = 0
|
COI = 12
12S = 0
16S = 1
18S = 0
28S = 0
|
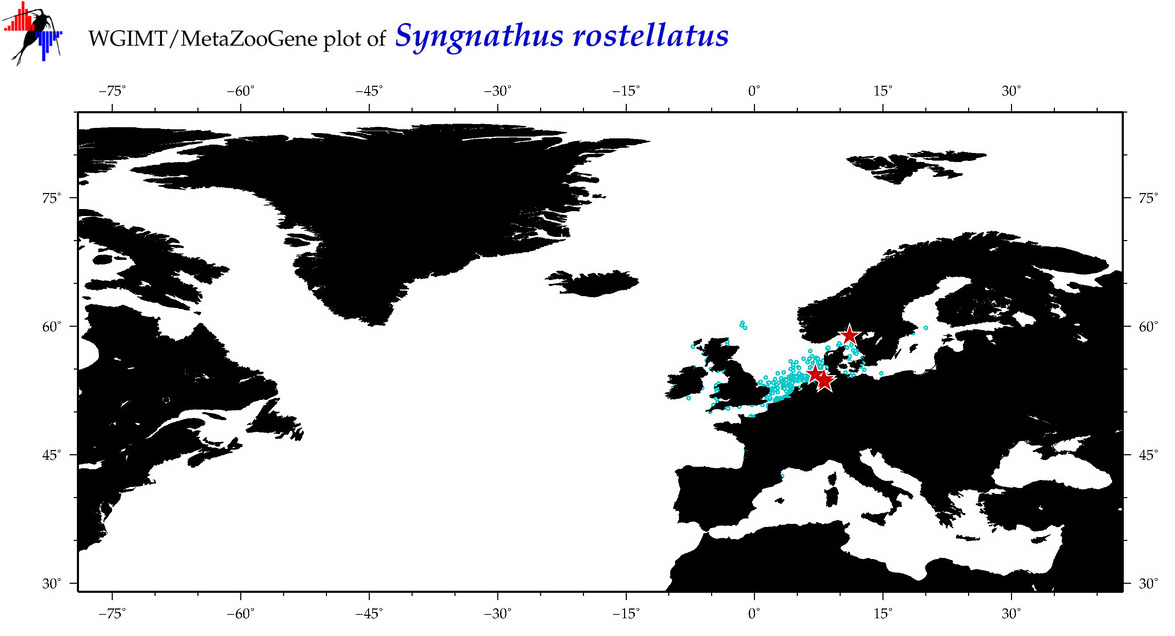 |
accepted
T5005680
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus scovelli |
Species
(51) |
COI
12S
16S
18S
28S
|
COI = 13
12S = 4
16S = 4
18S = 0
28S = 0
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005682
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus springeri |
Species
(52) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026072
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus typhle |
Species
(53) |
COI
12S
16S
18S
28S
|
COI = 49
12S = 8
16S = 8
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 1
18S = 0
28S = 0
|
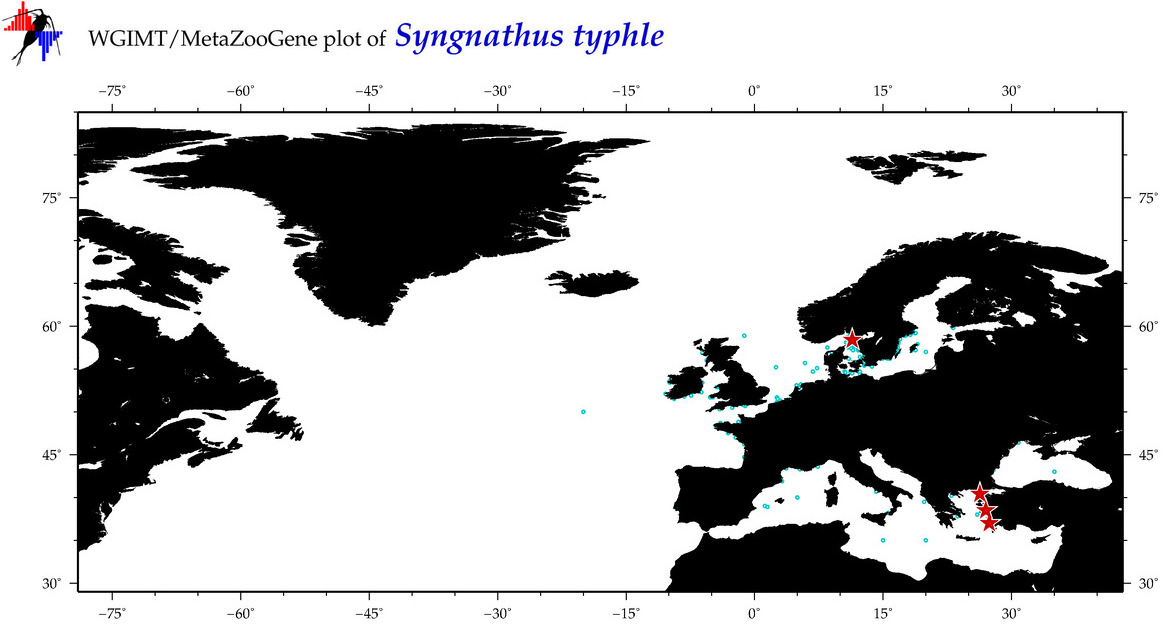 |
accepted
T5005683
R:1:1:0:0 |