Taxa Name
| Taxa
Level | Barcode
Data? | Number of
Barcodes
from
any ocean | Number of
Barcodes
from this region | Barcode Locations and
Species Occurence Map | MZGdb Atlas
v2026-m01-01
2026-Jan-26 |
| Syngnathiformes |
Order |
cummulative
sub-totals
------->
for this
Order |
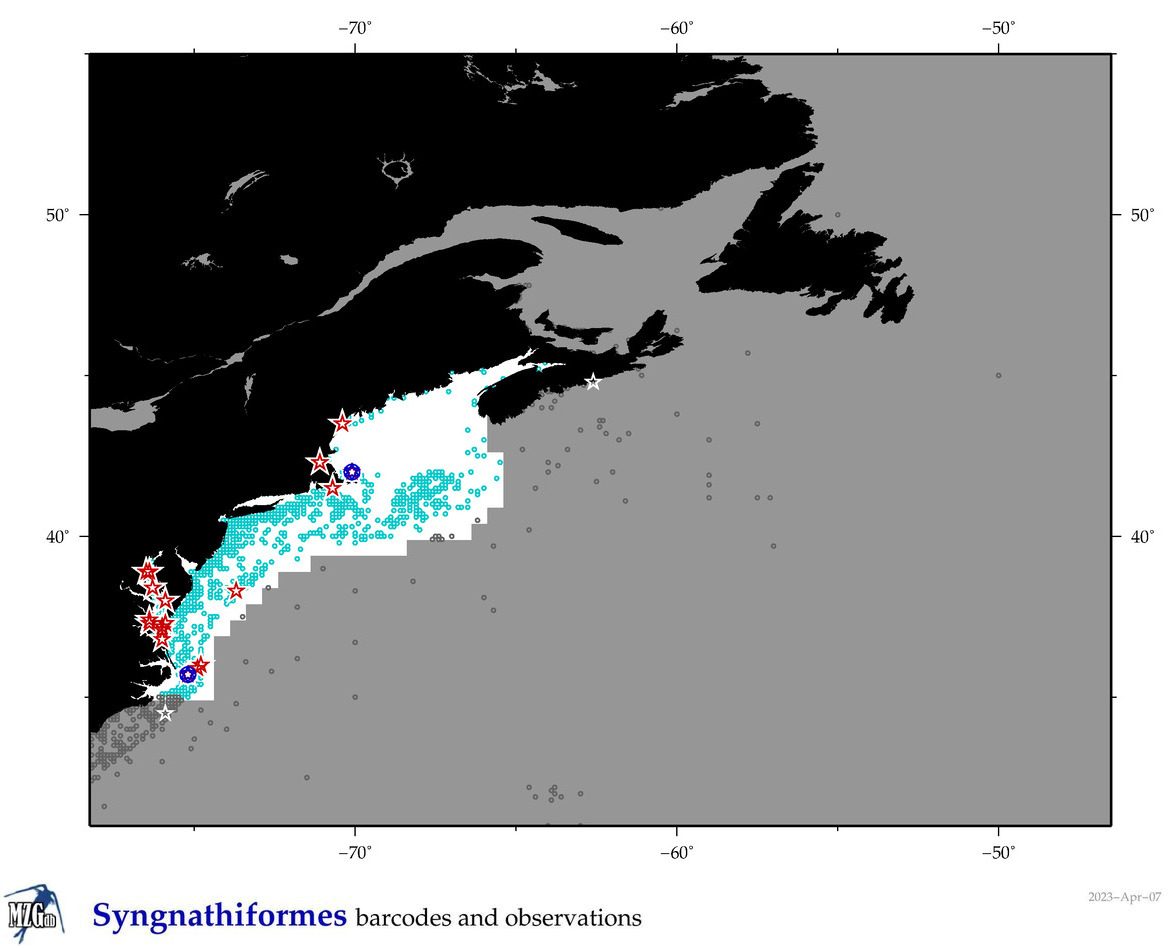
Total Species: 13
Sp. w/COI-Barcodes: 13
Total COI-Barcodes: 483 |
Total Species: 13
Sp. w/COI-Barcodes: 8
Total COI-Barcodes: 42 |
 |
accepted
T5002224
R:1:1:1:0 |
| ⋄ Aulostomidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 14 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5000237
R:1:0:0:0 |
| ⋄ ⋄ Aulostomus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 14 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002165
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Aulostomus maculatus |
Species
(1) |
COI
12S
16S
18S
28S
|
COI = 14
12S = 2
16S = 4
18S = 1
28S = 1
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5002166
R:1:0:0:0 |
| ⋄ Centriscidae |
Family |
cummulative
sub-totals
------->
for this
Family |
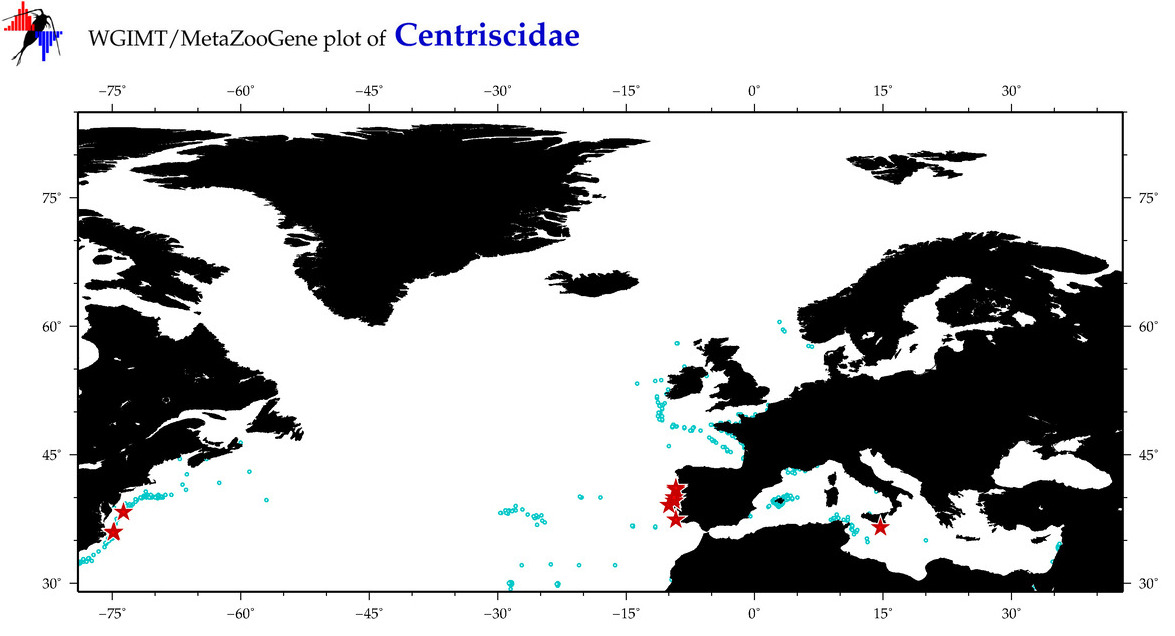
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 104 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 3 |
 |
accepted
T5000355
R:1:1:0:0 |
| ⋄ ⋄ Macroramphosinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
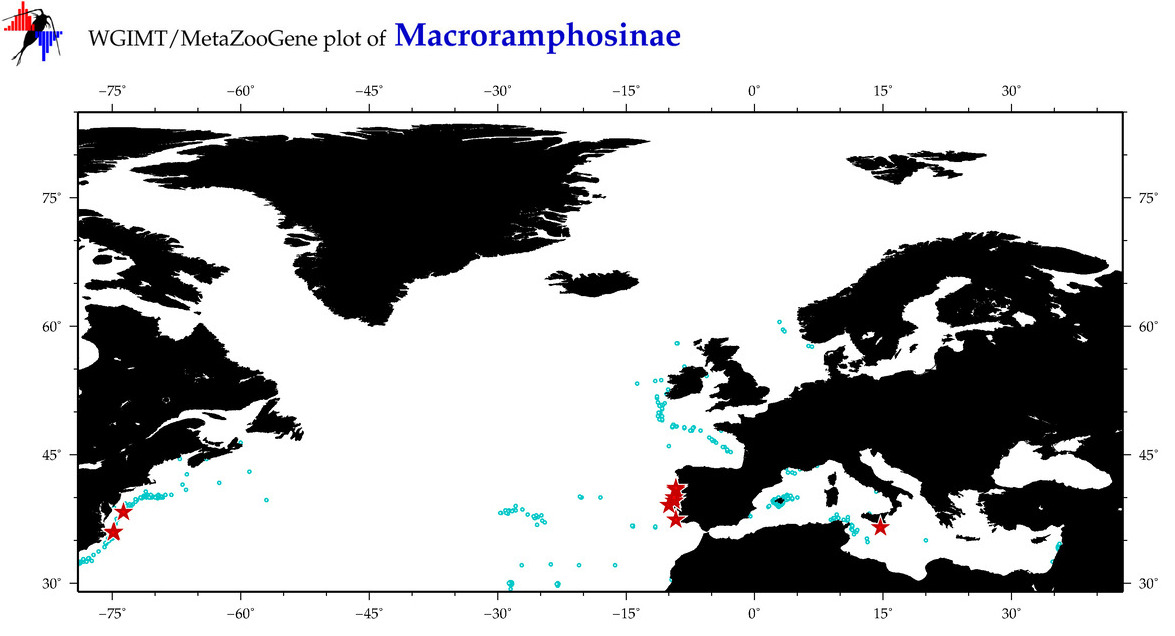
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 104 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 3 |
 |
accepted
T5002355
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Macroramphosus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 104 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 3 |
 |
accepted
T5000857
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Macroramphosus gracilis |
Species
(2) |
COI
12S
16S
18S
28S
|
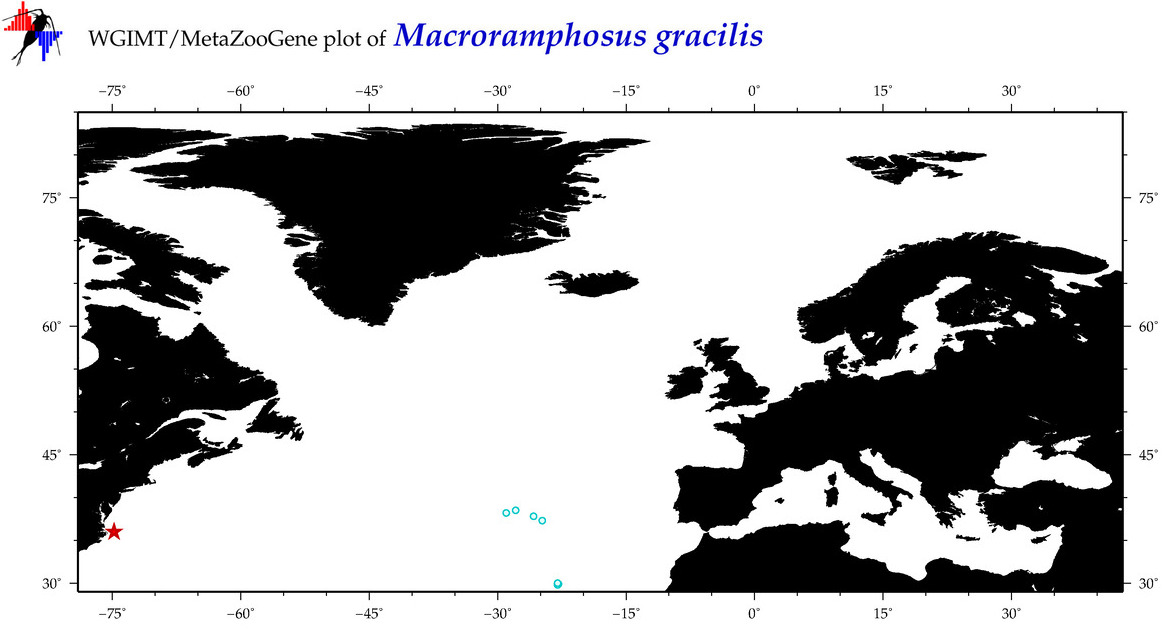
COI = 17
12S = 5
16S = 4
18S = 0
28S = 0
|
COI = 1
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000856
R:1:0:0:0 |
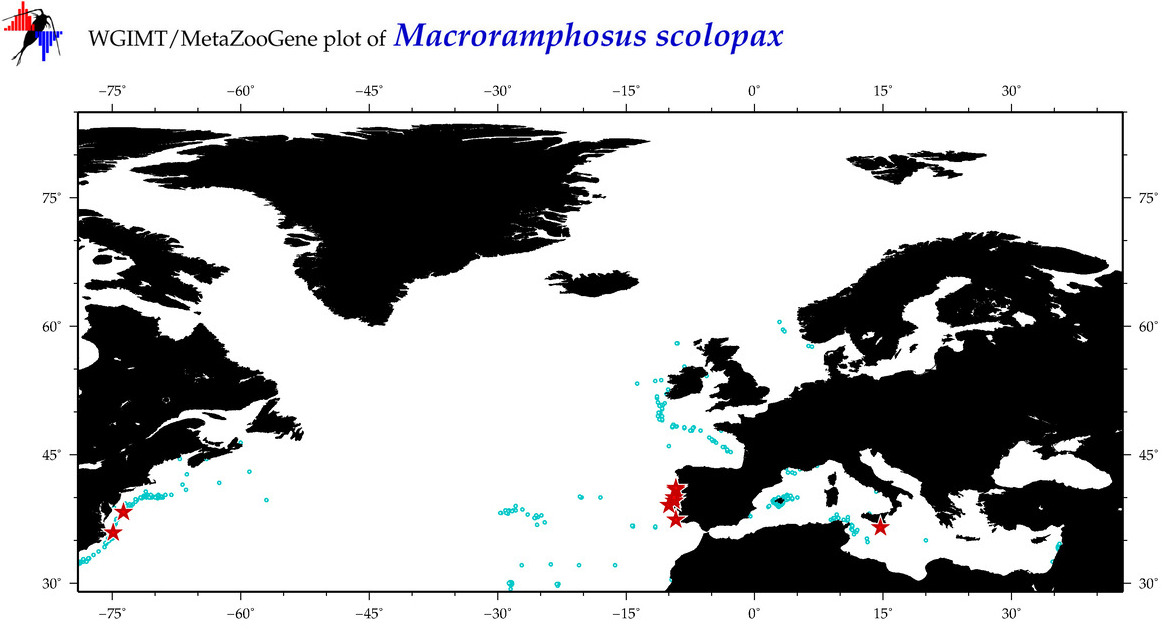
| ⋄ ⋄ ⋄ ⋄ Macroramphosus scolopax |
Species
(3) |
COI
12S
16S
18S
28S
|
COI = 87
12S = 6
16S = 20
18S = 0
28S = 1
|
COI = 2
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5000858
R:1:0:0:0 |
| ⋄ Fistulariidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 55 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 4 |
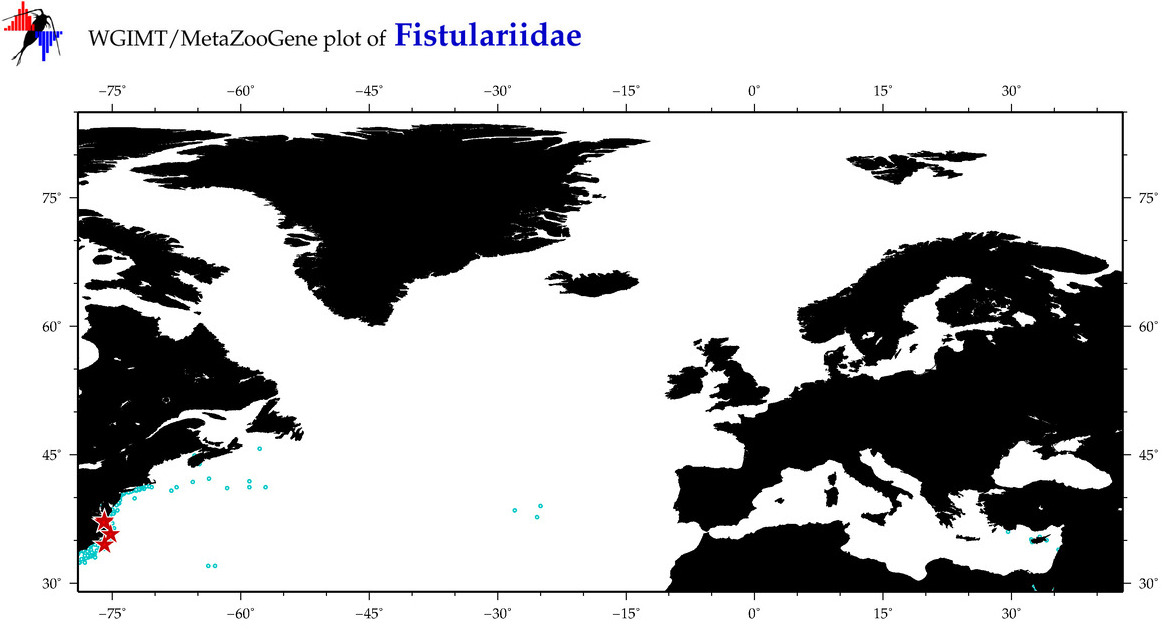 |
accepted
T5000584
R:1:1:0:0 |
| ⋄ ⋄ Fistularia |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 55 |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 4 |
 |
accepted
T5000583
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Fistularia petimba |
Species
(4) |
COI
12S
16S
18S
28S
|
COI = 45
12S = 15
16S = 11
18S = 1
28S = 0
|
COI = 1
12S = 1
16S = 1
18S = 1
28S = 0
|
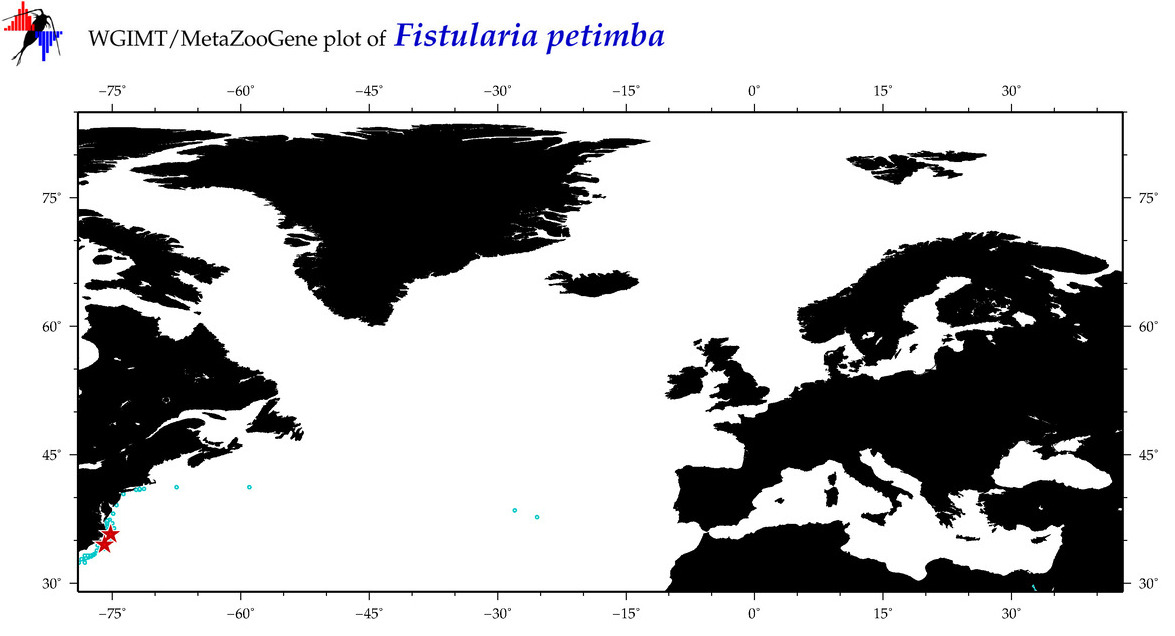 |
accepted
T5003998
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Fistularia tabacaria |
Species
(5) |
COI
12S
16S
18S
28S
|
COI = 10
12S = 2
16S = 2
18S = 0
28S = 0
|
COI = 3
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003999
R:1:1:0:0 |
| ⋄ Syngnathidae |
Family |
cummulative
sub-totals
------->
for this
Family |
Total Species: 8
Sp. w/COI-Barcodes: 8
Total COI-Barcodes: 310 |
Total Species: 8
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 35 |
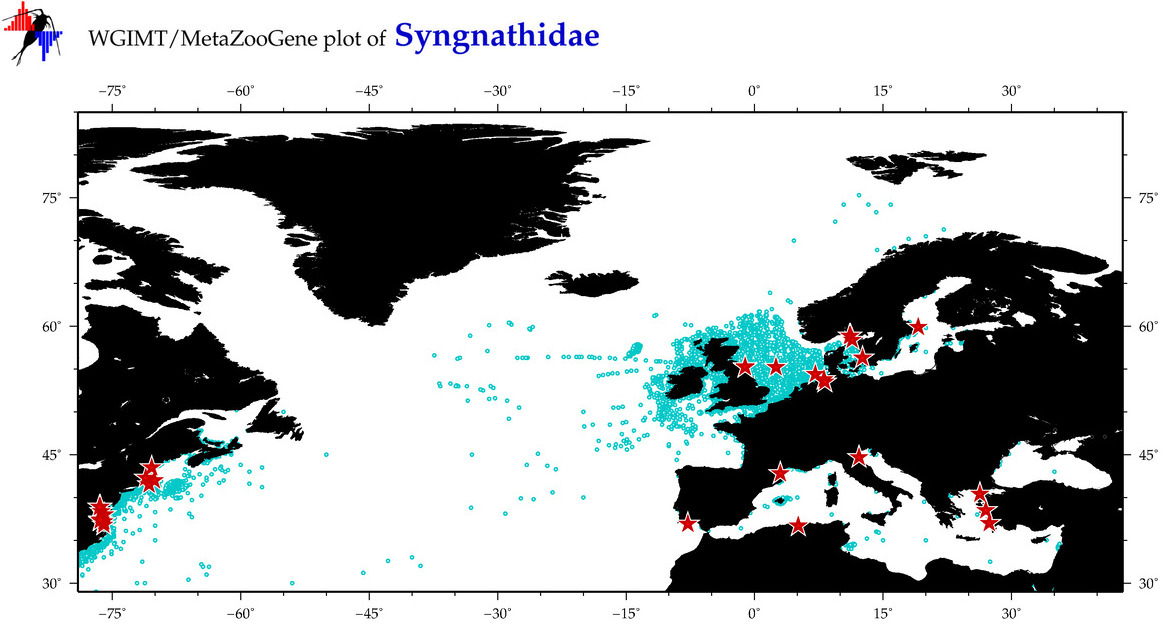 |
accepted
T5000121
R:1:1:1:0 |
| ⋄ ⋄ Hippocampinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 103 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
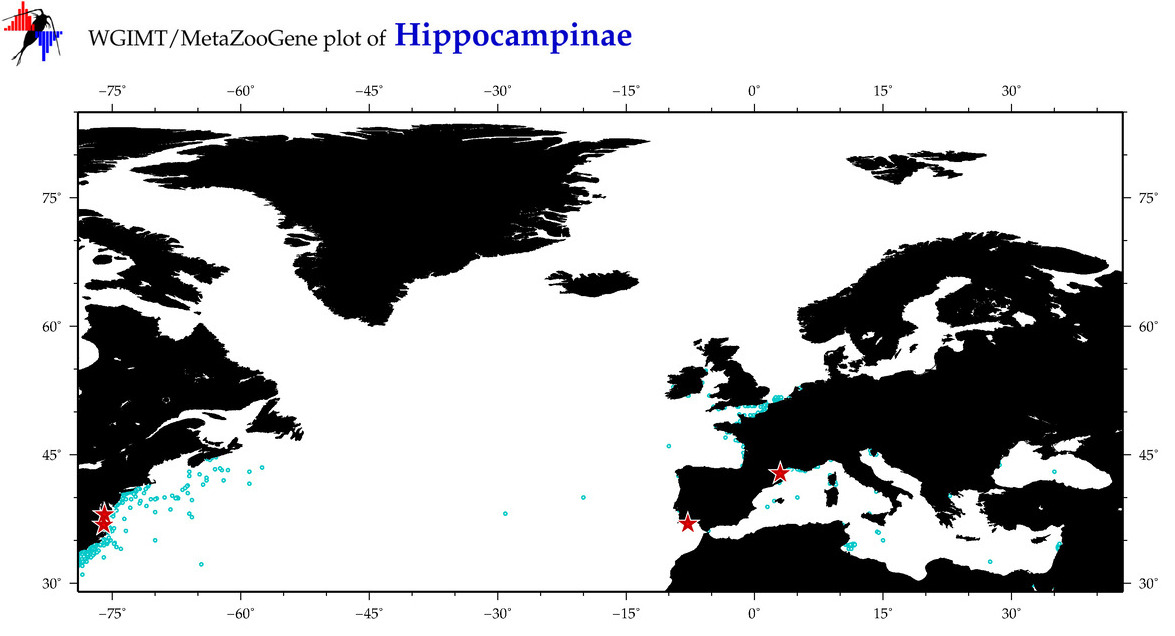 |
accepted
T5002357
R:1:1:0:0 |
| ⋄ ⋄ ⋄ Hippocampus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 2
Sp. w/COI-Barcodes: 2
Total COI-Barcodes: 103 |
Total Species: 2
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 4 |
 |
accepted
T5000668
R:1:1:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus erectus |
Species
(6) |
COI
12S
16S
18S
28S
|
COI = 49
12S = 5
16S = 18
18S = 0
28S = 0
|
COI = 4
12S = 0
16S = 0
18S = 0
28S = 0
|
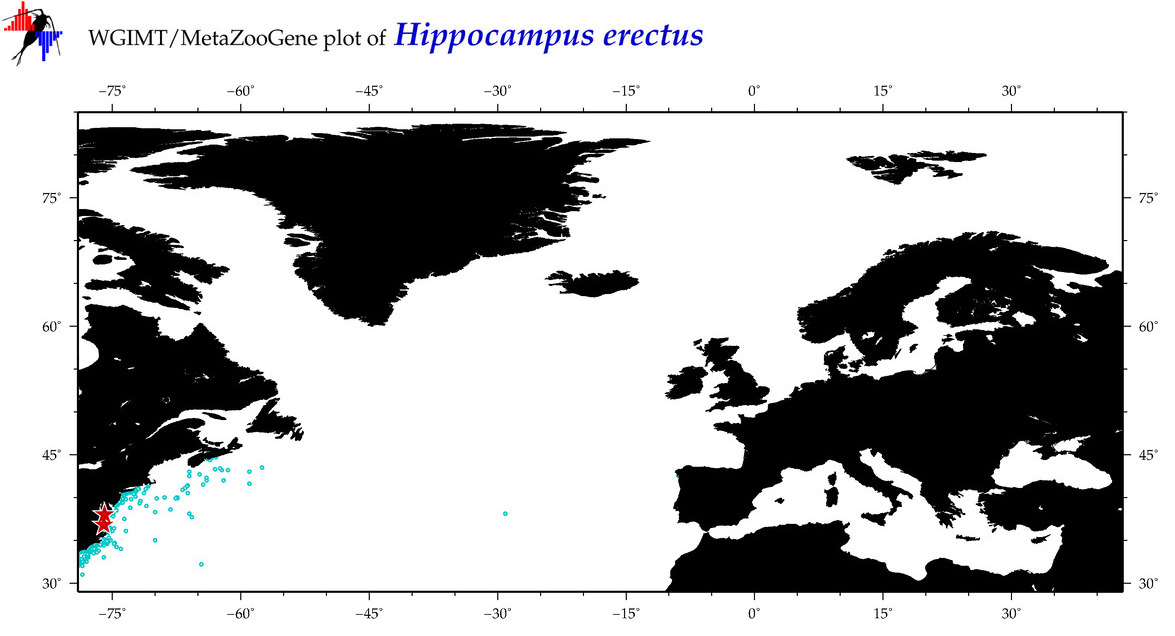 |
accepted
T5000669
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Hippocampus reidi |
Species
(7) |
COI
12S
16S
18S
28S
|
COI = 54
12S = 4
16S = 9
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5004274
R:1:1:0:0 |
| ⋄ ⋄ Syngnathinae |
Subfamily |
cummulative
sub-totals
------->
for this
Subfamily |
Total Species: 6
Sp. w/COI-Barcodes: 6
Total COI-Barcodes: 207 |
Total Species: 6
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 31 |
 |
accepted
T5002356
R:1:1:1:0 |
| ⋄ ⋄ ⋄ Amphelikturus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 3 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
 |
accepted
T5002440
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Amphelikturus dendriticus |
Species
(8) |
COI
12S
16S
18S
28S
|
COI = 3
12S = 1
16S = 1
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5003086
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Phyllopteryx |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 1
Sp. w/COI-Barcodes: 1
Total COI-Barcodes: 144 |
Total Species: 1
Sp. w/COI-Barcodes: 0
Total COI-Barcodes: 0 |
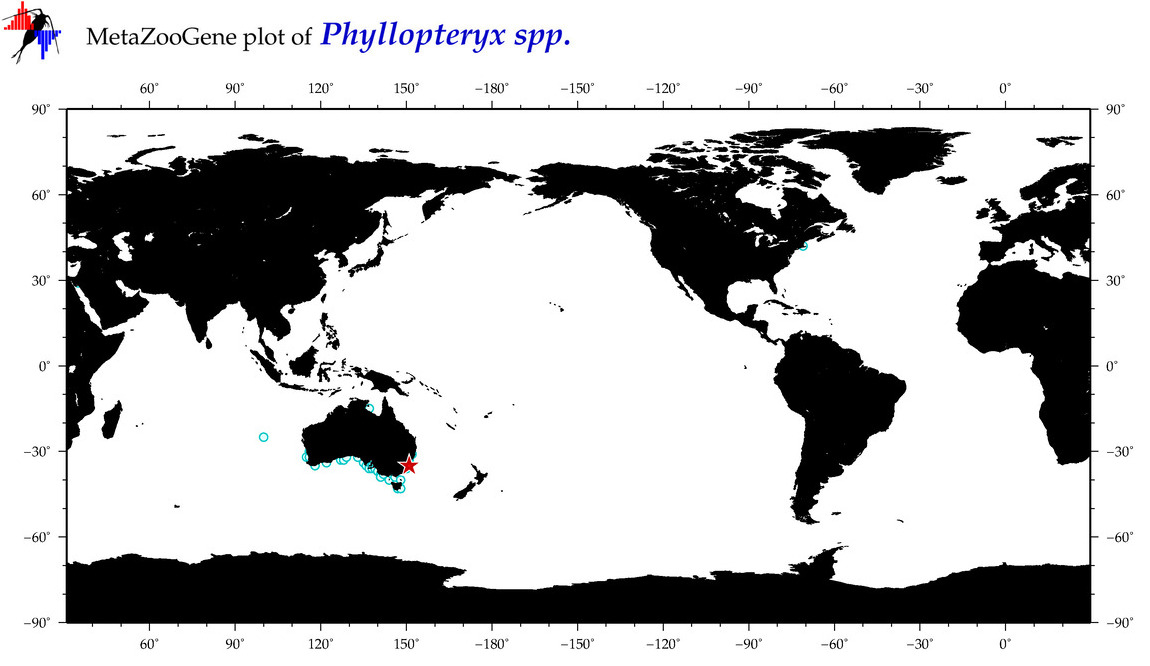 |
accepted
T5002855
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Phyllopteryx taeniolatus |
Species
(9) |
COI
12S
16S
18S
28S
|
COI = 144
12S = 8
16S = 8
18S = 1
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
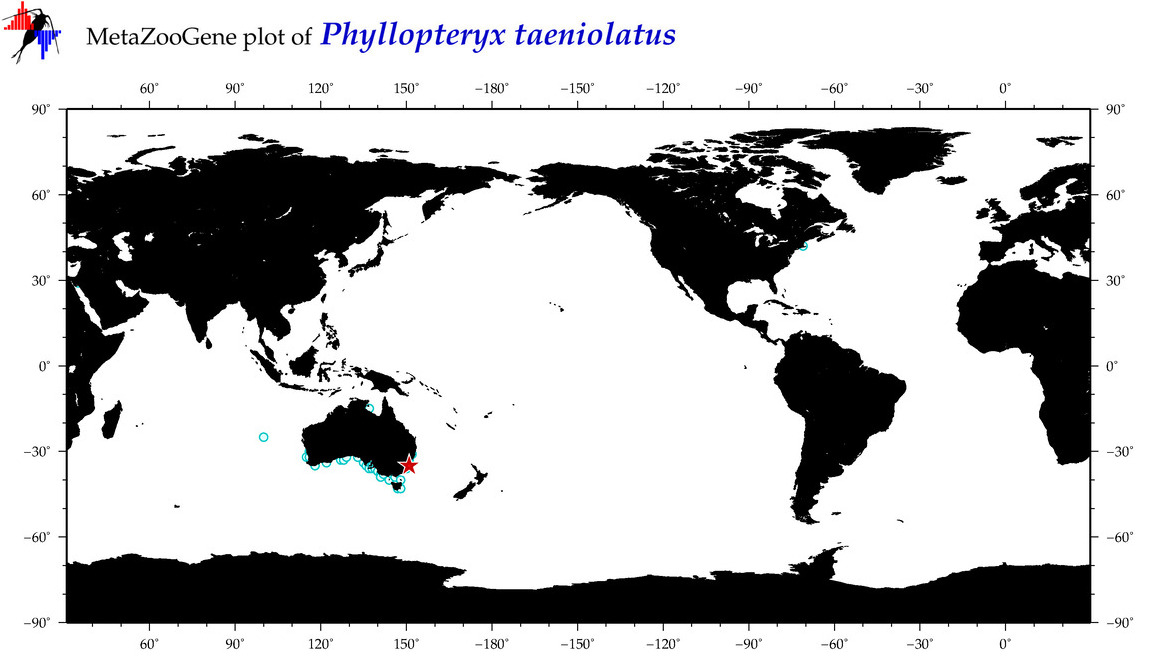 |
accepted
T5005040
R:1:0:0:0 |
| ⋄ ⋄ ⋄ Syngnathus |
Genus |
cummulative
sub-totals
------->
for this
Genus |
Total Species: 4
Sp. w/COI-Barcodes: 4
Total COI-Barcodes: 60 |
Total Species: 4
Sp. w/COI-Barcodes: 3
Total COI-Barcodes: 31 |
 |
accepted
T5001322
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus floridae |
Species
(10) |
COI
12S
16S
18S
28S
|
COI = 20
12S = 5
16S = 5
18S = 0
28S = 0
|
COI = 8
12S = 0
16S = 0
18S = 0
28S = 0
|
 |
accepted
T5005678
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus fuscus |
Species
(11) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 3
16S = 2
18S = 1
28S = 0
|
COI = 16
12S = 1
16S = 1
18S = 1
28S = 0
|
 |
accepted
T5005679
R:1:1:1:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus louisianae |
Species
(12) |
COI
12S
16S
18S
28S
|
COI = 18
12S = 3
16S = 3
18S = 1
28S = 0
|
COI = 7
12S = 0
16S = 0
18S = 0
28S = 0
|
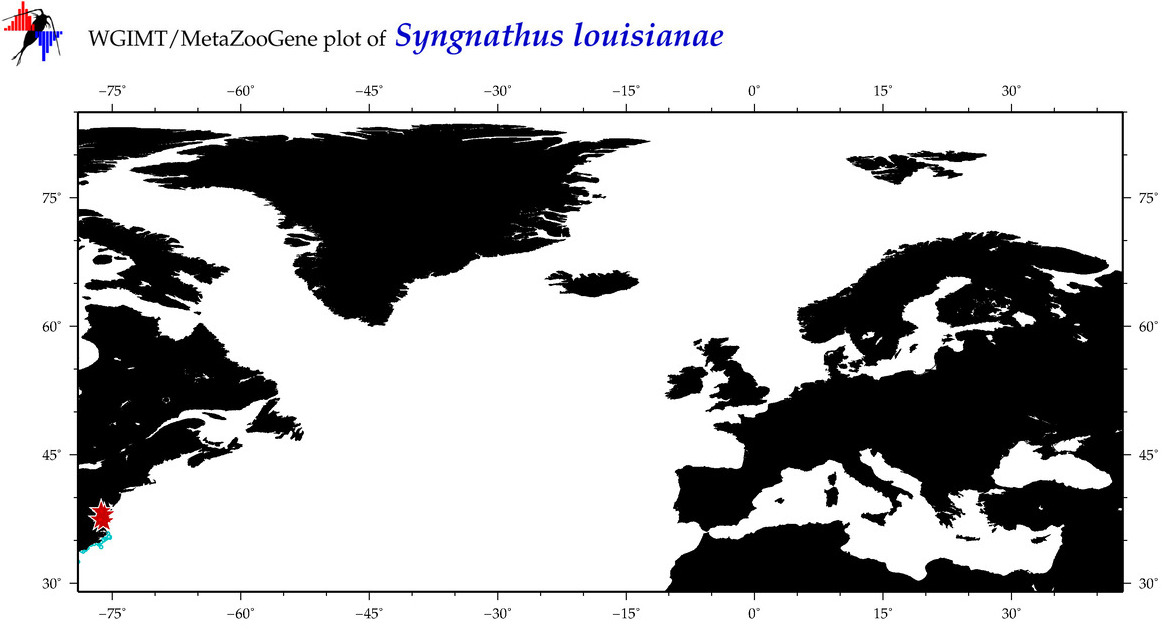 |
accepted
T5001325
R:1:0:0:0 |
| ⋄ ⋄ ⋄ ⋄ Syngnathus springeri |
Species
(13) |
COI
12S
16S
18S
28S
|
COI = 4
12S = 1
16S = 1
18S = 0
28S = 0
|
COI = 0
12S = 0
16S = 0
18S = 0
28S = 0
|
no map |
accepted
T5026072
R:1:0:0:0 |